
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,267,028 – 14,267,294 |
| Length | 266 |
| Max. P | 0.983174 |

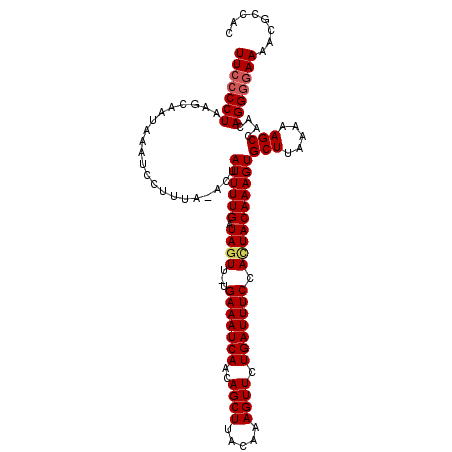
| Location | 14,267,028 – 14,267,139 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 91.99 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -20.83 |
| Energy contribution | -21.45 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14267028 111 + 22224390 UUCCCCUAAGCAAUAAAUCCUUUA-ACUAUUUUGAUAGUU-UGAAAUCAACAGCUUACAAAGUUCUGAUUUCCAGUACAAAGUGCUCAAGAAGCCAAUAGGGGAAAACGCCAC ((((((((.((............(-(((((....))))))-.(((((((..((((.....)))).))))))).(((((...)))))......))...))))))))........ ( -24.90) >DroSec_CAF1 7690 112 + 1 UUCCCCUAAGCAAUAAAUCCUUUAAAUUAUUUUGAUAGUU-UGAAAUCAACAGCUUAAAAAGUUCUGAUUUCCACUACAAAGUGCUUAAAAAGCCAACAGGGGAAAACGCCAC (((((((.....................((((((.((((.-.(((((((..((((.....)))).))))))).))))))))))(((.....)))....)))))))........ ( -25.80) >DroSim_CAF1 11681 112 + 1 UUCCCCUAAGCAAUAAAUCCUUUAAACUAUUUUGAUAGUU-UGAAAUCAACAGCUUACAAAGUUCUGAUUUCCACUACAAAGUGCUUAAAAAGCCAACAGGGGAAAACGCCAC (((((((.....................((((((.((((.-.(((((((..((((.....)))).))))))).))))))))))(((.....)))....)))))))........ ( -25.80) >DroYak_CAF1 10863 112 + 1 UUCGCCUAAGUAAUAAGCCCUUUA-AGUAUUUUGAUAGUUUUGAAAUCAACAGCUUGCAAAGUUCUGAUUUCCAUUACAAAGUGCUUAAGAAGCCAACAGGGAAAAACAUCAC (((.(((..((.....((..((((-(((((((((.((((...(((((((..((((.....)))).))))))).)))))))))))))))))..))..))))))))......... ( -24.10) >consensus UUCCCCUAAGCAAUAAAUCCUUUA_ACUAUUUUGAUAGUU_UGAAAUCAACAGCUUACAAAGUUCUGAUUUCCACUACAAAGUGCUUAAAAAGCCAACAGGGGAAAACGCCAC (((((((.....................((((((.((((...(((((((..((((.....)))).))))))).))))))))))(((.....)))....)))))))........ (-20.83 = -21.45 + 0.62)

| Location | 14,267,099 – 14,267,214 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.39 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -33.14 |
| Energy contribution | -32.42 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14267099 115 - 22224390 GAGA-----GAUGUGUUUAAGGAUGAUUGAUUGCGAGUCUUAACUCAAUGGGCGGCUUGACAUCGAUAAAUUUGCGAUAGGUGGCGUUUUCCCCUAUUGGCUUCUUGAGCACUUUGUACU ....-----...(((((((((((..((((((..((((((....((.....)).))))))..)))))).......(((((((.((......)))))))))..)))))))))))........ ( -32.10) >DroSec_CAF1 7762 115 - 1 GGGA-----GAUGUGUUUAAGGAUGAUUGAUUGCGAGCCUUAACUCAAUGGGCGGCUUGACAUCGAUAAAUUUCCGAUAGGUGGCGUUUUCCCCUGUUGGCUUUUUAAGCACUUUGUAGU ....-----...(((((((((((..((((((..((((((....((.....)).))))))..))))))......((((((((.((......)))))))))).)))))))))))........ ( -34.50) >DroSim_CAF1 11753 115 - 1 GGGA-----GAUGUGUUUAAGGAUGAUUGAUUGCGAGCCUUAACUCAAUGGGCGGCUUGACAUCGAUAAAUUUCCGAUAGGUGGCGUUUUCCCCUGUUGGCUUUUUAAGCACUUUGUAGU ....-----...(((((((((((..((((((..((((((....((.....)).))))))..))))))......((((((((.((......)))))))))).)))))))))))........ ( -34.50) >DroEre_CAF1 10662 115 - 1 GAGA-----GAUGUGUUUAAGGAUGAUUGAUUGCGAGCCUUAACUCAAUGGGCGGCUUGACAUCGAUAAAUUUCCGAUAGGUGGCGUUCUCCCCUGUUGGCUUCUUAAACACUUUGUAAU ....-----...(((((((((((..((((((..((((((....((.....)).))))))..))))))......((((((((.((......)))))))))).)))))))))))........ ( -37.00) >DroYak_CAF1 10935 120 - 1 UGGAGAGAGGAUGUGUUUAAGGAUGAUUGAUUGCAAGGCUUAACUCAAUGGGCGGCUUGACAUCGAUAAAUUUGCGAUAGGUGAUGUUUUUCCCUGUUGGCUUCUUAAGCACUUUGUAAU ............(((((((((((..((((((..(((((((((......)))))..))))..)))))).......(((((((.((......)))))))))..)))))))))))........ ( -32.20) >consensus GGGA_____GAUGUGUUUAAGGAUGAUUGAUUGCGAGCCUUAACUCAAUGGGCGGCUUGACAUCGAUAAAUUUCCGAUAGGUGGCGUUUUCCCCUGUUGGCUUCUUAAGCACUUUGUAAU ............(((((((((((..((((((..((((((....((.....)).))))))..))))))......((((((((.((......)))))))))).)))))))))))........ (-33.14 = -32.42 + -0.72)


| Location | 14,267,179 – 14,267,294 |
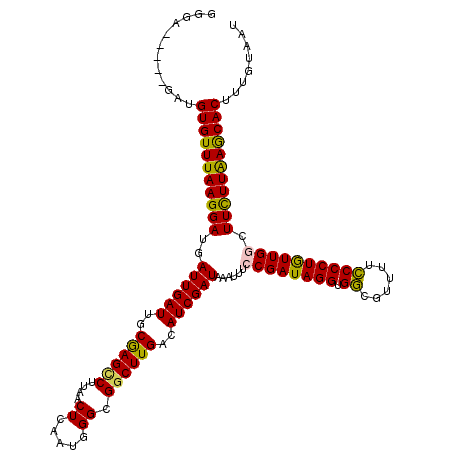
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.96 |
| Mean single sequence MFE | -39.02 |
| Consensus MFE | -33.92 |
| Energy contribution | -34.04 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.926281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14267179 115 - 22224390 AUGCCGGAAAAGUCCCGGGGCCCGGAGAUUUUGUUUGAGCCUGAUUGGCAAGAUGGCUGGCAGUCGAUUGGACCUGAAUCGAGA-----GAUGUGUUUAAGGAUGAUUGAUUGCGAGUCU ....((((((((((((((...)))).)))))).)))).(((.....))).....((((.(((((((((((..(((..(((....-----))).......))).))))))))))).)))). ( -40.30) >DroSec_CAF1 7842 115 - 1 AUGCCGGAAAAGUCCCGGGGCCCGGAGAUUUUGCUUGAGCCUGAUUGGCAAGAUGGUUGGCAGUCGAUUGGACCUGAAUCGGGA-----GAUGUGUUUAAGGAUGAUUGAUUGCGAGCCU .((((((.((((((((((...)))).))))))((....))....))))))....((((.(((((((((((..(((..(((....-----))).......))).))))))))))).)))). ( -41.50) >DroSim_CAF1 11833 115 - 1 AUGCCGGAAAAGUCCCGGGGCCCAGAGAUUUUGCUUGAGCCUGAUUGGCAAGAUGGUUGGCAGUCGAUUGGACCUGAAUCGGGA-----GAUGUGUUUAAGGAUGAUUGAUUGCGAGCCU .(((((((....)))..((((...(((......)))..))))....))))....((((.(((((((((((..(((..(((....-----))).......))).))))))))))).)))). ( -39.20) >DroEre_CAF1 10742 115 - 1 AUGCCGGAAAAGUCCCGGGGCCCGGAGAUUUUGCUUGAGCCUGAUUGGCAAGAUGCCUGGCAGUCGAUUGGACCUGAAUCGAGA-----GAUGUGUUUAAGGAUGAUUGAUUGCGAGCCU ...((((.......))))(((.(((..((((((((...........)))))))).))..(((((((((((..(((..(((....-----))).......))).)))))))))))).))). ( -38.00) >DroYak_CAF1 11015 120 - 1 AUGCCGGAAAAGUCCCGGGGCCCGGAGAUUUUGCUUGAGCCUGAUUGGCAAGAUGGUUGGCAGUCGAUUGGACCUGAAUCUGGAGAGAGGAUGUGUUUAAGGAUGAUUGAUUGCAAGGCU ..(((((.((((((((((...)))).)))))).))...(((.....)))..........(((((((((((..(((..((((.......)))).......))).)))))))))))..))). ( -36.10) >consensus AUGCCGGAAAAGUCCCGGGGCCCGGAGAUUUUGCUUGAGCCUGAUUGGCAAGAUGGUUGGCAGUCGAUUGGACCUGAAUCGGGA_____GAUGUGUUUAAGGAUGAUUGAUUGCGAGCCU .((((((.((((((((((...)))).))))))((....))....))))))....((((.(((((((((((..(((..(((.........))).......))).))))))))))).)))). (-33.92 = -34.04 + 0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:25 2006