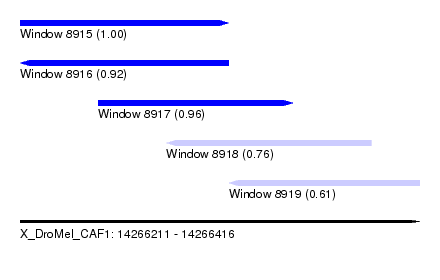
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,266,211 – 14,266,416 |
| Length | 205 |
| Max. P | 0.999844 |

| Location | 14,266,211 – 14,266,318 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 97.57 |
| Mean single sequence MFE | -29.14 |
| Consensus MFE | -27.04 |
| Energy contribution | -27.04 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.23 |
| SVM RNA-class probability | 0.999844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14266211 107 + 22224390 UCCAUCUUCGCAGCCAGCUGGCUGGCAAUGGAAAAACGUUUUUGCAACAGAAUAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAAGGCAAAUACA (((((...(.(((((....))))))..))))).....((.(((((..........(((((((((((..((((((....)))))).)))))))))))..))))).)). ( -30.50) >DroSec_CAF1 6893 107 + 1 UCCAUCUUCGCAGCCAGCUGGCAGGCAAUGGAAAAACGUUUUUGCAACAGAGCAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAAGGCAAAUACA ............(((.(((.(((((.((((......))))))))).....)))..(((((((((((..((((((....)))))).))))))))))).)))....... ( -28.10) >DroSim_CAF1 10881 107 + 1 UCCAUCUUCGCAGCCAGCUGGCAGGCAAUGGAAAAACGUUUUUGCAACAGAAUAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAAGGCAAAUACA (((((..((...(((....))).))..))))).....((.(((((..........(((((((((((..((((((....)))))).)))))))))))..))))).)). ( -27.70) >DroEre_CAF1 9996 107 + 1 UCCAUCUUCGCAGCCAGCUGGCUGGCAAUGGAAAAGCGUUUUUGCAACAGAAUAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAAAGCAAAUACA (((((...(.(((((....))))))..))))).....((.(((((..........(((((((((((..((((((....)))))).)))))))))))..))))).)). ( -29.40) >DroYak_CAF1 10119 107 + 1 UCCAUCUUCGCAGCCAGCUGGCAGGCAAUGGAAAAACGUUUUUGCAGCAGAAUAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAAGGCAAAUACA ............(((.((((.((((.((((......)))))))))))).......(((((((((((..((((((....)))))).))))))))))).)))....... ( -30.00) >consensus UCCAUCUUCGCAGCCAGCUGGCAGGCAAUGGAAAAACGUUUUUGCAACAGAAUAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAAGGCAAAUACA (((((..((...(((....))).))..))))).....((.(((((..........(((((((((((..((((((....)))))).)))))))))))..))))).)). (-27.04 = -27.04 + -0.00)


| Location | 14,266,211 – 14,266,318 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 97.57 |
| Mean single sequence MFE | -24.76 |
| Consensus MFE | -22.06 |
| Energy contribution | -22.30 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14266211 107 - 22224390 UGUAUUUGCCUUAAAUCACUCAUACGCAACGUUUUGCAUUUUGAAUAAUUUAUUAUUCUGUUGCAAAAACGUUUUUCCAUUGCCAGCCAGCUGGCUGCGAAGAUGGA ...................((((.((((((((((((((....((((((....))))))...))).))))))))........(((((....))))).)))...)))). ( -28.00) >DroSec_CAF1 6893 107 - 1 UGUAUUUGCCUUAAAUCACUCAUACGCAACGUUUUGCAUUUUGAAUAAUUUAUUGCUCUGUUGCAAAAACGUUUUUCCAUUGCCUGCCAGCUGGCUGCGAAGAUGGA ...........................(((((((((((....((.(((....))).))...))).))))))))..((((((((..(((....))).))...)))))) ( -20.60) >DroSim_CAF1 10881 107 - 1 UGUAUUUGCCUUAAAUCACUCAUACGCAACGUUUUGCAUUUUGAAUAAUUUAUUAUUCUGUUGCAAAAACGUUUUUCCAUUGCCUGCCAGCUGGCUGCGAAGAUGGA ...........................(((((((((((....((((((....))))))...))).))))))))..((((((((..(((....))).))...)))))) ( -24.60) >DroEre_CAF1 9996 107 - 1 UGUAUUUGCUUUAAAUCACUCAUACGCAACGUUUUGCAUUUUGAAUAAUUUAUUAUUCUGUUGCAAAAACGCUUUUCCAUUGCCAGCCAGCUGGCUGCGAAGAUGGA ...................((((.((((.(((((((((....((((((....))))))...))).))))))..........(((((....)))))))))...)))). ( -26.70) >DroYak_CAF1 10119 107 - 1 UGUAUUUGCCUUAAAUCACUCAUACGCAACGUUUUGCAUUUUGAAUAAUUUAUUAUUCUGCUGCAAAAACGUUUUUCCAUUGCCUGCCAGCUGGCUGCGAAGAUGGA ...........................(((((((((((....((((((....))))))...))).))))))))..((((((((..(((....))).))...)))))) ( -23.90) >consensus UGUAUUUGCCUUAAAUCACUCAUACGCAACGUUUUGCAUUUUGAAUAAUUUAUUAUUCUGUUGCAAAAACGUUUUUCCAUUGCCUGCCAGCUGGCUGCGAAGAUGGA ...........................(((((((((((....((((((....))))))...))).))))))))..((((((((..(((....))).))...)))))) (-22.06 = -22.30 + 0.24)


| Location | 14,266,251 – 14,266,351 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 87.09 |
| Mean single sequence MFE | -20.85 |
| Consensus MFE | -14.82 |
| Energy contribution | -14.83 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14266251 100 + 22224390 UUUGCAACAGAAUAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAAGGCAAAUACAAAUACUAUAUGUAUA----UGUAUCCACUUACAUAGA ...............(((((((((((..((((((....)))))).))))))))))).((...(((((.((((.....)))).----)))))))........... ( -21.10) >DroSim_CAF1 10921 100 + 1 UUUGCAACAGAAUAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAAGGCAAAUACAAAUACUAUAUGUAUA----UGUAUCCACAUACAUAGA ...............(((((((((((..((((((....)))))).))))))))))).((...(((((.((((.....)))).----)))))))........... ( -21.10) >DroEre_CAF1 10036 88 + 1 UUUGCAACAGAAUAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAAAGCAAAUACACAUACUAUACAUGUA----------------CAUAUA (((((..........(((((((((((..((((((....)))))).)))))))))))..)))))...((((.......)))).----------------...... ( -14.30) >DroYak_CAF1 10159 104 + 1 UUUGCAGCAGAAUAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAAGGCAAAUACAAAUACUAUAUAUGUAUGUAUGUAUGUACAUGCAUAGA (((((..........(((((((((((..((((((....)))))).)))))))))))..))))).............(((((((((((....))))))))))).. ( -26.90) >consensus UUUGCAACAGAAUAAUAAAUUAUUCAAAAUGCAAAACGUUGCGUAUGAGUGAUUUAAGGCAAAUACAAAUACUAUAUAUAUA____UGUAUCCACAUACAUAGA (((((..........(((((((((((..((((((....)))))).)))))))))))..)))))......................................... (-14.82 = -14.83 + 0.00)


| Location | 14,266,286 – 14,266,391 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 85.83 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -15.79 |
| Energy contribution | -16.10 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14266286 105 - 22224390 UGGUUACGCAUUUUUCGAUGCGAAUAUAUUUGCGAAUGUGUCUAUGUAAGUGGAUACA----UAUACAUAUAGUAUUUGUAUUUGCCUUAAAUCACUCAUACGCAACGU ......((((((....)))))).......(((((.(((.((.....((((..((((((----.((((.....)))).))))))..).)))....)).))).)))))... ( -28.20) >DroSim_CAF1 10956 105 - 1 UGGUUACGCAUUUUUCGAUGCGAAUAUAUUUGCGAAUGUGUCUAUGUAUGUGGAUACA----UAUACAUAUAGUAUUUGUAUUUGCCUUAAAUCACUCAUACGCAACGU ......((((((....)))))).......(((((.(((.((........(..((((((----.((((.....)))).))))))..)........)).))).)))))... ( -27.19) >DroEre_CAF1 10071 93 - 1 UGGGCACGCAUUUUCCGAUGCGAAUAUAUUUGCGAACGUGUAUAUG----------------UACAUGUAUAGUAUGUGUAUUUGCUUUAAAUCACUCAUACGCAACGU ......((((((....)))))).......((((..(((((((....----------------)))))))...(((((.(((((((...))))).)).)))))))))... ( -22.80) >DroYak_CAF1 10194 109 - 1 UGGUUACGCAUUUUUCCAUGCGAAUAUAUUUGCGAAUGUGUCUAUGCAUGUACAUACAUACAUACAUAUAUAGUAUUUGUAUUUGCCUUAAAUCACUCAUACGCAACGU .(((..(((((......)))))((((((..(((..((((((.((((.((((....)))).)))).)))))).)))..)))))).)))...................... ( -22.20) >consensus UGGUUACGCAUUUUUCGAUGCGAAUAUAUUUGCGAAUGUGUCUAUGUAUGUGGAUACA____UACACAUAUAGUAUUUGUAUUUGCCUUAAAUCACUCAUACGCAACGU ...................(((((((((..(((..((((((...(((((....))))).....))))))...)))..)))))))))....................... (-15.79 = -16.10 + 0.31)


| Location | 14,266,318 – 14,266,416 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.98 |
| Mean single sequence MFE | -22.39 |
| Consensus MFE | -12.61 |
| Energy contribution | -13.18 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14266318 98 - 22224390 GAAUAU----------AGCUCAUACAUAUC-----ACAUAUGGUUACGCAUUUUUCGAUGCGAAUAUAUUUGCGAAUGUGUCUAUGUAAGUGGAUACA----UAUACAUAUAGUAUU ((((((----------(.......(((((.-----..)))))....((((((....))))))..)))))))....((((((((((....)))))))))----)((((.....)))). ( -21.50) >DroSim_CAF1 10988 98 - 1 GGAUAU----------AGCUCAUACAUAUC-----CCAUAUGGUUACGCAUUUUUCGAUGCGAAUAUAUUUGCGAAUGUGUCUAUGUAUGUGGAUACA----UAUACAUAUAGUAUU ((((((----------(..((...(((((.-----..)))))....((((((....))))))...........)).)))))))(((((((((....))----)))))))........ ( -24.30) >DroEre_CAF1 10103 91 - 1 CGAUAU----------AGCUCAUACAUAUCCCAUCCCAUAUGGGCACGCAUUUUCCGAUGCGAAUAUAUUUGCGAACGUGUAUAUG----------------UACAUGUAUAGUAUG .(((((----------((((((((..............))))))).((((((....))))))..))))))(((..(((((((....----------------)))))))...))).. ( -22.54) >DroYak_CAF1 10226 112 - 1 GGAUAUAUACUCGUACAGCUCAUACAUAUC-----CCAUAUGGUUACGCAUUUUUCCAUGCGAAUAUAUUUGCGAAUGUGUCUAUGCAUGUACAUACAUACAUACAUAUAUAGUAUU ((((((((..(((((.(((.((((......-----...))))))).(((((......)))))........)))))))))))))(((.(((((......))))).))).......... ( -21.20) >consensus GGAUAU__________AGCUCAUACAUAUC_____CCAUAUGGUUACGCAUUUUUCGAUGCGAAUAUAUUUGCGAAUGUGUCUAUGUAUGUGGAUACA____UACACAUAUAGUAUU .................((..((((((((........)))))....((((((....))))))....)))..))..((((((...(((((....))))).....))))))........ (-12.61 = -13.18 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:22 2006