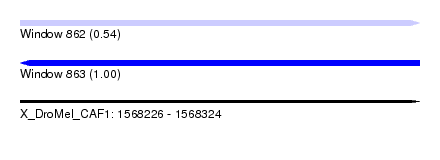
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,568,226 – 1,568,324 |
| Length | 98 |
| Max. P | 0.999731 |

| Location | 1,568,226 – 1,568,324 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.59 |
| Mean single sequence MFE | -36.17 |
| Consensus MFE | -14.99 |
| Energy contribution | -16.37 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1568226 98 + 22224390 UGCAGACUCCGGAAUCC-------AAAAUCCGGAAUCCGGAC-------UGAGGACUUUGGCUGUUAGCUGUUAGCUGUCAACUAAAAGCCCUCAGCCAGAG----CGAGCCUUGU ..(((..((((((.(((-------.......))).)))))))-------))(((.(((((((((...(((.((((.......)))).)))...)))))))))----....)))... ( -37.80) >DroSec_CAF1 29779 98 + 1 UGCAGACUCCGGAAUCC-------AAAAUCCGGAAUCCGGAC-------UGAGGACUUUGGCUGUUAGCUGUUAGCUGUCAACUAAAAGCCCUCAGCCAGAG----CGAGCCUUGC .((....((((((.(((-------.......))).)))))).-------.((((.(((((((((...(((.((((.......)))).)))...)))))))))----....)))))) ( -37.90) >DroEre_CAF1 33270 112 + 1 UGCAGAGUCCGGAAUGCAAAAUGCAAAAUCCGGAAUCCGGAAUCCGGACUGCGGACUUUACCAGUUAGCUGUUAGCUGUCAACUAAAAGCCCUCAGCCUGAG----AGAGCCUUGC .((((((((((((.(((.....)))...(((((...))))).))))))))..((.((((..(((...((((...(((..........)))...)))))))..----)))))))))) ( -40.50) >DroYak_CAF1 30255 102 + 1 UGCAGACUCCGGAAUCC-------AAAUUCCGAACUGAGGACUGAGGAUUGCGGACUCUCCUAGUUA-------GCUGUCAACUAAAAGCCCUCAGCCCGAAAGAAAGAGCCUUGC .((((.(((((((((..-------..))))))..((..((.((((((...((((.....))(((((.-------......)))))...)))))))).))...))...)))..)))) ( -28.50) >consensus UGCAGACUCCGGAAUCC_______AAAAUCCGGAAUCCGGAC_______UGAGGACUUUGCCAGUUAGCUGUUAGCUGUCAACUAAAAGCCCUCAGCCAGAG____AGAGCCUUGC .((((..((((((...............))))))..................((.((((........((((...(((..........)))...)))).........)))))))))) (-14.99 = -16.37 + 1.38)

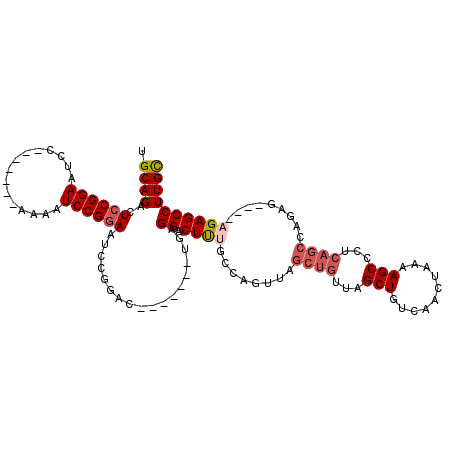
| Location | 1,568,226 – 1,568,324 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.59 |
| Mean single sequence MFE | -44.00 |
| Consensus MFE | -25.09 |
| Energy contribution | -27.27 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -4.24 |
| Structure conservation index | 0.57 |
| SVM decision value | 3.96 |
| SVM RNA-class probability | 0.999731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
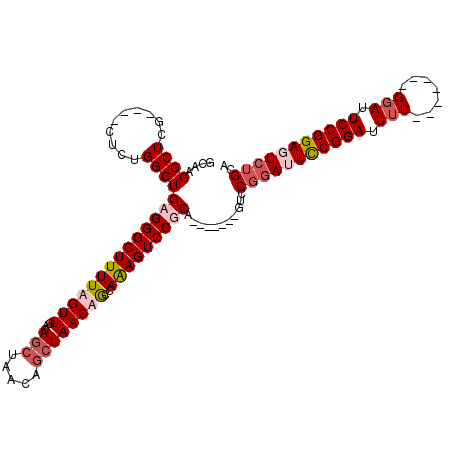
>X_DroMel_CAF1 1568226 98 - 22224390 ACAAGGCUCG----CUCUGGCUGAGGGCUUUUAGUUGACAGCUAACAGCUAACAGCCAAAGUCCUCA-------GUCCGGAUUCCGGAUUUU-------GGAUUCCGGAGUCUGCA .........(----(...(((((((((((((..((((..(((.....)))..)))).))))))))))-------))).((((((((((....-------....)))))))))))). ( -44.40) >DroSec_CAF1 29779 98 - 1 GCAAGGCUCG----CUCUGGCUGAGGGCUUUUAGUUGACAGCUAACAGCUAACAGCCAAAGUCCUCA-------GUCCGGAUUCCGGAUUUU-------GGAUUCCGGAGUCUGCA (((.(((((.----....(((((((((((((..((((..(((.....)))..)))).))))))))))-------)))((((.((((.....)-------))).)))))))))))). ( -47.20) >DroEre_CAF1 33270 112 - 1 GCAAGGCUCU----CUCAGGCUGAGGGCUUUUAGUUGACAGCUAACAGCUAACUGGUAAAGUCCGCAGUCCGGAUUCCGGAUUCCGGAUUUUGCAUUUUGCAUUCCGGACUCUGCA ((((((((((----(.......))))))))(((((((........)))))))((((.((((((((.(((((((...))))))).))))))))((.....))...))))....))). ( -42.70) >DroYak_CAF1 30255 102 - 1 GCAAGGCUCUUUCUUUCGGGCUGAGGGCUUUUAGUUGACAGC-------UAACUAGGAGAGUCCGCAAUCCUCAGUCCUCAGUUCGGAAUUU-------GGAUUCCGGAGUCUGCA (((.((((((.......((((((((((((..((((((.....-------))))))(((..(....)..)))..))))))))))))(((((..-------..)))))))))))))). ( -41.70) >consensus GCAAGGCUCG____CUCUGGCUGAGGGCUUUUAGUUGACAGCUAACAGCUAACAGCCAAAGUCCGCA_______GUCCGGAUUCCGGAUUUU_______GGAUUCCGGAGUCUGCA ....((((..........))))((((((((((((((...(((.....))))))))..)))))))))...........(((((((((((.(((.......))).))))))))))).. (-25.09 = -27.27 + 2.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:28 2006