
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,225,146 – 14,225,246 |
| Length | 100 |
| Max. P | 0.599065 |

| Location | 14,225,146 – 14,225,246 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 90.27 |
| Mean single sequence MFE | -22.93 |
| Consensus MFE | -17.99 |
| Energy contribution | -19.05 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14225146 100 + 22224390 UACAUGAUGCAUUUUAUAUUUUGCAAAUGCAUUACGUAUACGCCGUGUAUUCGCGAUGCAAUCACGCCGAAAAUUCGUUUCCAUAUAUUACAC--AAG------------CGAU (((.((((((((((((.....)).)))))))))).)))..(((.(((((...(((.((....)))))(((....)))...........)))))--..)------------)).. ( -21.70) >DroSec_CAF1 133744 112 + 1 UGCAUGAUGCAUUUUAUAUUUUGCAAAUGCAUUACGUAUACGCCGUGUAUUCGCGAUGCAAUCACGCCGAAAAUUCGAUUCCAUAUAUUACAC--AAGCACACCUACACGCGAU (((.((((((((((((.....)).)))))))))).)))..(((.(((((...(((.((....)))))(((....)))................--.........)))))))).. ( -23.00) >DroEre_CAF1 121431 114 + 1 UGCAUGAUGCAUUUUAUAUUUUACAAAUGCAUUACGUAUACGCCGUGUAUCCGCGAUGCAAUCACGCCGAAAAUUCGUUUCCAUAUAUUACACACAGCCACACGUACACGCGAU (((.((((((((((((.....)).)))))))))).)))..(((.((((((..(((.((....)))))(((....)))..........................))))))))).. ( -23.50) >DroYak_CAF1 124229 112 + 1 UGCAUGAUGCAUUUUAUAUUUUACAAAUGCAUUACGUAUACGCCGUGUAUUCGCGAUGCAAUCACGCCGAAAAUUCGUUUCCAUAUAUUACAC--UAGCACACGUACACGCGAU (((.((((((((((((.....)).)))))))))).)))..(((.((((((..(((.((....)))))(((....)))................--........))))))))).. ( -23.50) >consensus UGCAUGAUGCAUUUUAUAUUUUACAAAUGCAUUACGUAUACGCCGUGUAUUCGCGAUGCAAUCACGCCGAAAAUUCGUUUCCAUAUAUUACAC__AAGCACACGUACACGCGAU (((.((((((((((((.....)).)))))))))).)))..(((.(((((...(((.((....)))))(((....)))...........................)))))))).. (-17.99 = -19.05 + 1.06)

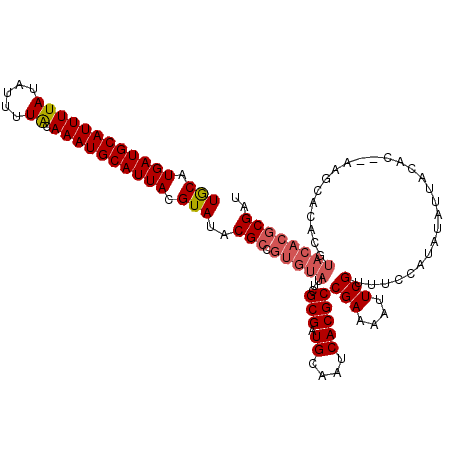
| Location | 14,225,146 – 14,225,246 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 90.27 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -22.74 |
| Energy contribution | -26.05 |
| Covariance contribution | 3.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14225146 100 - 22224390 AUCG------------CUU--GUGUAAUAUAUGGAAACGAAUUUUCGGCGUGAUUGCAUCGCGAAUACACGGCGUAUACGUAAUGCAUUUGCAAAAUAUAAAAUGCAUCAUGUA ..((------------((.--(((((.....((....)).........((((((...))))))..)))))))))..(((((.((((((((..........)))))))).))))) ( -27.60) >DroSec_CAF1 133744 112 - 1 AUCGCGUGUAGGUGUGCUU--GUGUAAUAUAUGGAAUCGAAUUUUCGGCGUGAUUGCAUCGCGAAUACACGGCGUAUACGUAAUGCAUUUGCAAAAUAUAAAAUGCAUCAUGCA ...(((((((.((((((((--(((((.....(((((......))))).((((((...))))))..))))))).)))))).))((((((((..........))))))))))))). ( -34.00) >DroEre_CAF1 121431 114 - 1 AUCGCGUGUACGUGUGGCUGUGUGUAAUAUAUGGAAACGAAUUUUCGGCGUGAUUGCAUCGCGGAUACACGGCGUAUACGUAAUGCAUUUGUAAAAUAUAAAAUGCAUCAUGCA ...((((((((((((((((((((((......((....)).........((((((...)))))).))))))))).))))))))((((((((..........))))))))))))). ( -41.80) >DroYak_CAF1 124229 112 - 1 AUCGCGUGUACGUGUGCUA--GUGUAAUAUAUGGAAACGAAUUUUCGGCGUGAUUGCAUCGCGAAUACACGGCGUAUACGUAAUGCAUUUGUAAAAUAUAAAAUGCAUCAUGCA ...((((((((((((((..--(((((.....((....)).........((((((...))))))..)))))...)))))))))((((((((..........))))))))))))). ( -36.60) >consensus AUCGCGUGUACGUGUGCUU__GUGUAAUAUAUGGAAACGAAUUUUCGGCGUGAUUGCAUCGCGAAUACACGGCGUAUACGUAAUGCAUUUGCAAAAUAUAAAAUGCAUCAUGCA ...((((((((((((((....(((((...........(((....))).((((((...))))))..)))))...)))))))))((((((((..........))))))))))))). (-22.74 = -26.05 + 3.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:46 2006