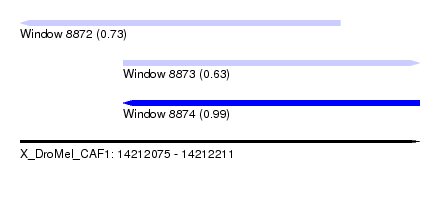
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,212,075 – 14,212,211 |
| Length | 136 |
| Max. P | 0.988886 |

| Location | 14,212,075 – 14,212,184 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -33.36 |
| Consensus MFE | -24.28 |
| Energy contribution | -24.24 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14212075 109 - 22224390 AUGAAA--ACACGUGGUUGACGCUUAUGACAGUGGGCCACUUUUGGCGCCACUCCUGGCCAAAAAUUCAUGCCCAGUGUUAACCAAACAAAGUUGCUGUGGCCAUUUGUCC ......--.....(((((((((((......(((((((((....)))).)))))...(((...........))).))))))))))).(((((...((....))..))))).. ( -32.70) >DroSec_CAF1 123231 111 - 1 AAAAACUUCCACGUGGUUGAACCUACUGGCAGUUGGCCACUUUUGGCGCCACUCCUGGCCAAAAAUUCAUGCCCAGUGUUAACCAAACAAAGUCGCUGCGGCCAUUUGUCC .............(((((((...((((((((.(((((((....((....))....))))))).......)).))))))))))))).(((((...((....))..))))).. ( -29.50) >DroSim_CAF1 99495 111 - 1 AAAAACUUCCACGUGGUUGAACCUACUGGCAGUUGGCCACUUUUGGCGCCACUCCUGGCCAAAAAUUCAUGCCCAGUGUUAACCAAACAAAGUCGCUGUGGCCAUUUGUCC ...((((.(((.((((......))))))).))))((((((....((((.(....((((.((........)).))))((((.....))))..).))))))))))........ ( -31.30) >DroEre_CAF1 110934 89 - 1 ----------------------GCCCAUGCAGUGGGCCGCUUUUGGCGCCAUUCCUGGCCAGAAAUCCAUGGCCAGUGUUGACCAAACAAAGUCGCUGUGGCCGCUUGUUC ----------------------((((((...)))))).......((((((((..(((((((........)))))))....(((........)))...)))).))))..... ( -31.80) >DroYak_CAF1 113183 111 - 1 AAAAACUUCCAUGUGGCUGGAGGUCAUGGCAGUGGGCCACUUUUGGCGCAACUCCUGGCCAGAAAUCCAUGGCCAGUGUUAACCAAACAAAGUCGCUGUGGCCAUUUGUCC ....(((((((......)))))))...(((((..((((((....(((((.....(((((((........)))))))((((.....))))..).))))))))))..))))). ( -41.50) >consensus AAAAACUUCCACGUGGUUGAACCUACUGGCAGUGGGCCACUUUUGGCGCCACUCCUGGCCAAAAAUUCAUGCCCAGUGUUAACCAAACAAAGUCGCUGUGGCCAUUUGUCC ...........................(((((..((((((...((((.......((((.((........)).))))((((.....))))..))))..))))))..))))). (-24.28 = -24.24 + -0.04)


| Location | 14,212,110 – 14,212,211 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 83.01 |
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -19.42 |
| Energy contribution | -21.80 |
| Covariance contribution | 2.38 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14212110 101 + 22224390 CUGGGCAUGAAUUUUUGGCCAGGAGUGGCGCCAAAAGUGGCCCACUGUCAUAAGCGUCAACCACGUGU--UUUCAUUUUUUCAUUGCCCUUCUUUUCUUUUGA ..(((((((((....((((....(((((.((((....)))))))))))))...((((.....))))..--.........)))).))))).............. ( -28.80) >DroSec_CAF1 123266 103 + 1 CUGGGCAUGAAUUUUUGGCCAGGAGUGGCGCCAAAAGUGGCCAACUGCCAGUAGGUUCAACCACGUGGAAGUUUUUUUUUUCAUUGCCCCUCUUUUAUUUUGA ..(((((((((((((((((.((...((((((.....)).)))).))))))).))))))).....(((((((......)))))))))))).............. ( -30.90) >DroSim_CAF1 99530 103 + 1 CUGGGCAUGAAUUUUUGGCCAGGAGUGGCGCCAAAAGUGGCCAACUGCCAGUAGGUUCAACCACGUGGAAGUUUUUUUUUUCAUUGCCCCUCUUUUAUUUUGA ..(((((((((((((((((.((...((((((.....)).)))).))))))).))))))).....(((((((......)))))))))))).............. ( -30.90) >DroYak_CAF1 113218 103 + 1 CUGGCCAUGGAUUUCUGGCCAGGAGUUGCGCCAAAAGUGGCCCACUGCCAUGACCUCCAGCCACAUGGAAGUUUUUCCCUUUACCCAUUCUUUUUUCUUUUGA (((((((........))))))).........((((((((((.....)))).(((.((((......)))).))).......................)))))). ( -26.70) >consensus CUGGGCAUGAAUUUUUGGCCAGGAGUGGCGCCAAAAGUGGCCAACUGCCAGUAGCUUCAACCACGUGGAAGUUUUUUUUUUCAUUGCCCCUCUUUUAUUUUGA ..(((((((((((((((((....(((.(.((((....))))).))))))).)))))))).....(((((((......)))))))))))).............. (-19.42 = -21.80 + 2.38)


| Location | 14,212,110 – 14,212,211 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.01 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -21.70 |
| Energy contribution | -21.82 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14212110 101 - 22224390 UCAAAAGAAAAGAAGGGCAAUGAAAAAAUGAAA--ACACGUGGUUGACGCUUAUGACAGUGGGCCACUUUUGGCGCCACUCCUGGCCAAAAAUUCAUGCCCAG ..............(((((.((((.........--....((((((.((.(....)...)).))))))(((((((..........))))))).))))))))).. ( -30.30) >DroSec_CAF1 123266 103 - 1 UCAAAAUAAAAGAGGGGCAAUGAAAAAAAAAACUUCCACGUGGUUGAACCUACUGGCAGUUGGCCACUUUUGGCGCCACUCCUGGCCAAAAAUUCAUGCCCAG ..............(((((.((((...............(((((..(.((....))...)..)))))(((((((..........))))))).))))))))).. ( -31.40) >DroSim_CAF1 99530 103 - 1 UCAAAAUAAAAGAGGGGCAAUGAAAAAAAAAACUUCCACGUGGUUGAACCUACUGGCAGUUGGCCACUUUUGGCGCCACUCCUGGCCAAAAAUUCAUGCCCAG ..............(((((.((((...............(((((..(.((....))...)..)))))(((((((..........))))))).))))))))).. ( -31.40) >DroYak_CAF1 113218 103 - 1 UCAAAAGAAAAAAGAAUGGGUAAAGGGAAAAACUUCCAUGUGGCUGGAGGUCAUGGCAGUGGGCCACUUUUGGCGCAACUCCUGGCCAGAAAUCCAUGGCCAG .................((((....((((....)))).((((.(..(((((...(((.....))))))))..)))))))))(((((((........))))))) ( -32.50) >consensus UCAAAAGAAAAGAGGGGCAAUGAAAAAAAAAACUUCCACGUGGUUGAACCUAAUGGCAGUGGGCCACUUUUGGCGCCACUCCUGGCCAAAAAUUCAUGCCCAG ..............(((((.((((...............(((((..(............)..)))))(((((((..........))))))).))))))))).. (-21.70 = -21.82 + 0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:42 2006