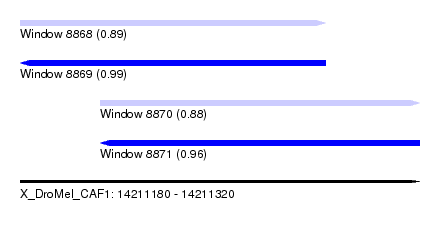
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,211,180 – 14,211,320 |
| Length | 140 |
| Max. P | 0.986506 |

| Location | 14,211,180 – 14,211,287 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 87.31 |
| Mean single sequence MFE | -35.54 |
| Consensus MFE | -25.92 |
| Energy contribution | -27.36 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14211180 107 + 22224390 UGAGCCUGGAGUC--UUC--UCCUCUUGGCUUAAAUGUGCAGCAUUGCAGAAAAUAUAUCGGCCAAAAUGCUGAGGCUGAGCUGCGGCCCAUCCUCCAUUUUGGCCCAUUC ((((((.((((..--..)--)))....))))))..(.(((......))).).........((((((((((..((((.((.((....)).)).))))))))))))))..... ( -40.50) >DroSec_CAF1 122356 109 + 1 UGAGCCUGGUGGUGUCUC--UCCUCUUGGCUUAAAUGUGCAGCAUUGCAGAAAAUAUAUCGGCCAAAAUGCUGAGGCUGAGCUGCGGCGCAUCCUCCAUUUUGGCCCAUUC ((((((.((.((......--.)).)).))))))..(.(((......))).).........((((((((((..((((.((.((....)).)).))))))))))))))..... ( -39.30) >DroSim_CAF1 98518 109 + 1 UGAGCCUGGUGGUGUCUC--UCCUCUUGGCUUAAAUGUGCAGCAUUGCAGAAAAUAUAUCGGCCAAAAUGCUGAGGCUGAGCUGCGGCGCAUCCUCCAUUUUGGCCCAUUC ((((((.((.((......--.)).)).))))))..(.(((......))).).........((((((((((..((((.((.((....)).)).))))))))))))))..... ( -39.30) >DroEre_CAF1 110031 96 + 1 ------------UGUCUC--UUCUCUUGGCUUAAAUGUGCAGCAUUGCAGAAAAUAUUUCGGCCAAAAUGCUGAGGCUGAGCUGCGGCCCAUCCUCCAUU-UGGACCAUUC ------------......--......(((..((((((..(((((((((.(((.....))).))...))))))).(((((.....))))).......))))-))..)))... ( -24.80) >DroYak_CAF1 112261 108 + 1 UGGGUUUCA---UGUCUCUCCUUUUUUGGCUUAAAUGUGCAGCAUUGCAGAAAAUAUUUCGGCCAAAAUGCUGAGGCUGAGCUGCGGCCCAUCCUGCAUUUUGGCCCAUUC (((((((((---.(((((.....((((((((.((((((((......))).....))))).))))))))....))))))))..(((((......)))))....))))))... ( -33.80) >consensus UGAGCCUGG_G_UGUCUC__UCCUCUUGGCUUAAAUGUGCAGCAUUGCAGAAAAUAUAUCGGCCAAAAUGCUGAGGCUGAGCUGCGGCCCAUCCUCCAUUUUGGCCCAUUC ((((((.....................))))))..(.(((......))).).........((((((((((..((((.((.((....)).)).))))))))))))))..... (-25.92 = -27.36 + 1.44)


| Location | 14,211,180 – 14,211,287 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 87.31 |
| Mean single sequence MFE | -35.36 |
| Consensus MFE | -28.78 |
| Energy contribution | -29.78 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14211180 107 - 22224390 GAAUGGGCCAAAAUGGAGGAUGGGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAUAUAUUUUCUGCAAUGCUGCACAUUUAAGCCAAGAGGA--GAA--GACUCCAGGCUCA (((((((((((((((((((.(((((....))))).))))..))))))))))...........((((....))))))))).((((....(((--(..--..)))).)))).. ( -42.20) >DroSec_CAF1 122356 109 - 1 GAAUGGGCCAAAAUGGAGGAUGCGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAUAUAUUUUCUGCAAUGCUGCACAUUUAAGCCAAGAGGA--GAGACACCACCAGGCUCA (((((((((((((((((((.((.((....)).)).))))..))))))))))...........((((....))))))))).((((..(.((.--......)).)..)))).. ( -35.50) >DroSim_CAF1 98518 109 - 1 GAAUGGGCCAAAAUGGAGGAUGCGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAUAUAUUUUCUGCAAUGCUGCACAUUUAAGCCAAGAGGA--GAGACACCACCAGGCUCA (((((((((((((((((((.((.((....)).)).))))..))))))))))...........((((....))))))))).((((..(.((.--......)).)..)))).. ( -35.50) >DroEre_CAF1 110031 96 - 1 GAAUGGUCCA-AAUGGAGGAUGGGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAAAUAUUUUCUGCAAUGCUGCACAUUUAAGCCAAGAGAA--GAGACA------------ ...((((..(-((((((((.(((((....))))).))))((((((...((.(((.....))).))))))))...)))))..))))......--......------------ ( -28.70) >DroYak_CAF1 112261 108 - 1 GAAUGGGCCAAAAUGCAGGAUGGGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAAAUAUUUUCUGCAAUGCUGCACAUUUAAGCCAAAAAAGGAGAGACA---UGAAACCCA ..(((((((((((((((((.(((((....))))).)))..))))))))))).......((((((((....))).........((......)))))))))---)........ ( -34.90) >consensus GAAUGGGCCAAAAUGGAGGAUGGGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAUAUAUUUUCUGCAAUGCUGCACAUUUAAGCCAAGAGGA__GAGACA_C_CCAGGCUCA (((((((((((((((((((.(((((....))))).))))..))))))))))...........((((....)))))))))................................ (-28.78 = -29.78 + 1.00)


| Location | 14,211,208 – 14,211,320 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.12 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -19.44 |
| Energy contribution | -22.32 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14211208 112 + 22224390 AAAUGUGCAGCAUUGCAGAAAAUAUAUCGGCCAAAAUGCUGAGGCUGAGCUGCGGCCCAUCCUCCAUUUUGGCCCAUUCACCUU--UUUGCCGGUCAGU--UUGGCAGUUG---GUAAU ...(.(((......))).).........((((((((((..((((.((.((....)).)).)))))))))))))).....(((..--.(((((((.....--)))))))..)---))... ( -38.50) >DroSec_CAF1 122386 89 + 1 AAAUGUGCAGCAUUGCAGAAAAUAUAUCGGCCAAAAUGCUGAGGCUGAGCUGCGGCGCAUCCUCCAUUUUGGCCCAUUCACC---------------------------UG---GUAAU ...(.(((......))).).........((((((((((..((((.((.((....)).)).))))))))))))))........---------------------------..---..... ( -27.70) >DroSim_CAF1 98548 112 + 1 AAAUGUGCAGCAUUGCAGAAAAUAUAUCGGCCAAAAUGCUGAGGCUGAGCUGCGGCGCAUCCUCCAUUUUGGCCCAUUCACCUU--UUUGCCGGUCAGU--UUGGCAGUUG---GUAAU ...(.(((......))).).........((((((((((..((((.((.((....)).)).)))))))))))))).....(((..--.(((((((.....--)))))))..)---))... ( -38.90) >DroEre_CAF1 110049 111 + 1 AAAUGUGCAGCAUUGCAGAAAAUAUUUCGGCCAAAAUGCUGAGGCUGAGCUGCGGCCCAUCCUCCAUU-UGGACCAUUCACCUU--UUUGCCGGUCAGU--UUGGCAGUUG---GUAAU ......((((((((......)))...((((((..........)))))))))))((.(((.........-))).))....(((..--.(((((((.....--)))))))..)---))... ( -32.80) >DroYak_CAF1 112290 112 + 1 AAAUGUGCAGCAUUGCAGAAAAUAUUUCGGCCAAAAUGCUGAGGCUGAGCUGCGGCCCAUCCUGCAUUUUGGCCCAUUCACCUU--UUUGCCGGUCACU--UUGGCAGUUG---GUAAU ...(.(((......))).).........(((((((((((.(((((((.....)))))..))..))))))))))).....(((..--.(((((((.....--)))))))..)---))... ( -40.30) >DroAna_CAF1 122419 101 + 1 AAAUGUGCAACAUUGCAGAAUAUAUUUCAAC----AUCAU---------C-CCCGCCCGGCCUCA----GACCUUAUUCACCUGACUCGGCUGGUCAGUUUUUGGCAGUUGAUGAUAAU .....((((....))))(((.....)))...----(((((---------(-...(.(((((((((----(...........))))...)))))).).((.....))....))))))... ( -21.60) >consensus AAAUGUGCAGCAUUGCAGAAAAUAUAUCGGCCAAAAUGCUGAGGCUGAGCUGCGGCCCAUCCUCCAUUUUGGCCCAUUCACCUU__UUUGCCGGUCAGU__UUGGCAGUUG___GUAAU ...(.(((......))).).........((((((((((..((((.((.((....)).)).)))))))))))))).............(((((((.......)))))))........... (-19.44 = -22.32 + 2.88)

| Location | 14,211,208 – 14,211,320 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.12 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -19.00 |
| Energy contribution | -21.58 |
| Covariance contribution | 2.59 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14211208 112 - 22224390 AUUAC---CAACUGCCAA--ACUGACCGGCAAA--AAGGUGAAUGGGCCAAAAUGGAGGAUGGGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAUAUAUUUUCUGCAAUGCUGCACAUUU .((((---(...((((..--.......))))..--..)))))((.((((((((((((((.(((((....))))).))))..)))))))))).))........((((....))))..... ( -40.50) >DroSec_CAF1 122386 89 - 1 AUUAC---CA---------------------------GGUGAAUGGGCCAAAAUGGAGGAUGCGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAUAUAUUUUCUGCAAUGCUGCACAUUU .....---..---------------------------...(((((((((((((((((((.((.((....)).)).))))..))))))))))...........((((....))))))))) ( -29.40) >DroSim_CAF1 98548 112 - 1 AUUAC---CAACUGCCAA--ACUGACCGGCAAA--AAGGUGAAUGGGCCAAAAUGGAGGAUGCGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAUAUAUUUUCUGCAAUGCUGCACAUUU .((((---(...((((..--.......))))..--..)))))((.((((((((((((((.((.((....)).)).))))..)))))))))).))........((((....))))..... ( -36.90) >DroEre_CAF1 110049 111 - 1 AUUAC---CAACUGCCAA--ACUGACCGGCAAA--AAGGUGAAUGGUCCA-AAUGGAGGAUGGGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAAAUAUUUUCUGCAAUGCUGCACAUUU .((((---(...((((..--.......))))..--..))))).(((((.(-((((((((.(((((....))))).))))..))))).)))))..........((((....))))..... ( -33.10) >DroYak_CAF1 112290 112 - 1 AUUAC---CAACUGCCAA--AGUGACCGGCAAA--AAGGUGAAUGGGCCAAAAUGCAGGAUGGGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAAAUAUUUUCUGCAAUGCUGCACAUUU .((((---(...((((..--.......))))..--..)))))(((((((((((((((((.(((((....))))).)))..)))))))))))...........((((....))))))).. ( -40.40) >DroAna_CAF1 122419 101 - 1 AUUAUCAUCAACUGCCAAAAACUGACCAGCCGAGUCAGGUGAAUAAGGUC----UGAGGCCGGGCGGG-G---------AUGAU----GUUGAAAUAUAUUCUGCAAUGUUGCACAUUU ..(((((((..(((((.....((((((....).)))))........(((.----....))).))))).-)---------)))))----).............((((....))))..... ( -28.20) >consensus AUUAC___CAACUGCCAA__ACUGACCGGCAAA__AAGGUGAAUGGGCCAAAAUGGAGGAUGGGCCGCAGCUCAGCCUCAGCAUUUUGGCCGAAAUAUUUUCUGCAAUGCUGCACAUUU ............((((...........)))).........(((((((((((((((((((.((.((....)).)).))))..))))))))))...........((((....))))))))) (-19.00 = -21.58 + 2.59)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:40 2006