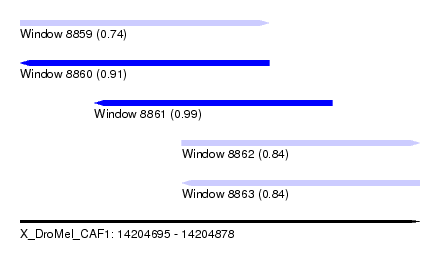
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,204,695 – 14,204,878 |
| Length | 183 |
| Max. P | 0.986102 |

| Location | 14,204,695 – 14,204,809 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 85.10 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -24.20 |
| Energy contribution | -26.07 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
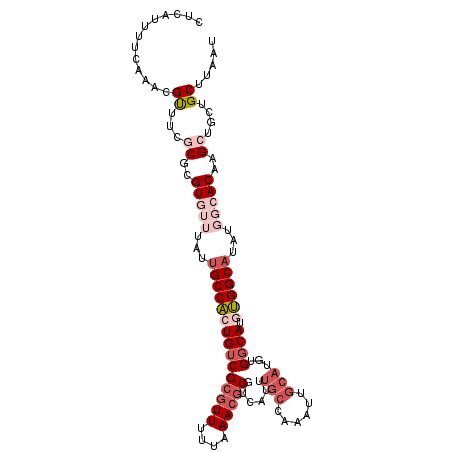
>X_DroMel_CAF1 14204695 114 + 22224390 UGAACCUUCUCUGCAGAUGGUUCGCUCUCAUUACUCCUCGUACAUGUCGCAUUUAGUUGGCUGCCGGCUGGCGCAUUAAGCAGCAGCUUGUGCCAUAUGCCACAUGCCACAUGC .(((((.(((....))).)))))((...((((((.....))).)))..)).....((.(((((..((((((((((...(((....))))))))))...))).)).))))).... ( -34.60) >DroSec_CAF1 116013 112 + 1 CGAACCUU--CUGCAGAUGGUUCGCUCUCAUUACUACUCGUACAUGUCGCAUUUAGUUGGCUAGCGGCUGGUGCAUUAAGCAGCAGCUUGUGCCAUAUGCCACAUGCCACAUGC ((((((.(--(....)).))))))...............((.(((((.((((.....((((....(((((.(((.....))).)))))...)))).)))).)))))..)).... ( -37.90) >DroSim_CAF1 92039 112 + 1 CGAACCUU--CUGCAGAUGGUUCGCUCUCAUUACUACUCGUACAUGUCGCAUUUAGUUGGCUGGCGGCUGGUGCAUUAAGCAGCAGCUUGUGCCAUAUGCCACAUGCCACAUGC ((((((.(--(....)).))))))..................(((((.((((...((.((((((((((((.(((.....))).)))))...))))...))))))))).))))). ( -39.90) >DroYak_CAF1 105976 96 + 1 UGGACCGU--CUGCAGAUAGUU------CAUUACUCCUCGUACAUGUCGCAUUUAGCUGACUGGCGGCUGGUGCAUUAAGCAG---CCAGU-------GCCGUAUGCCAUAUGC (((((..(--(....))..)))------))........(((((..(((((.....)).))).((((.(((((((.....))).---)))))-------))))))))........ ( -29.90) >consensus CGAACCUU__CUGCAGAUGGUUCGCUCUCAUUACUACUCGUACAUGUCGCAUUUAGUUGGCUGGCGGCUGGUGCAUUAAGCAGCAGCUUGUGCCAUAUGCCACAUGCCACAUGC ((((((............))))))..................(((((.((((...((((((....(((((.(((.....))).)))))...))))...))...)))).))))). (-24.20 = -26.07 + 1.87)

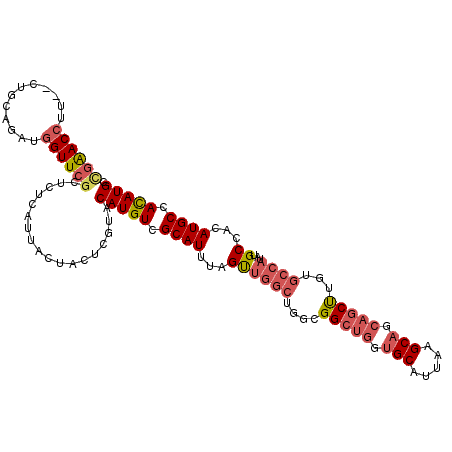
| Location | 14,204,695 – 14,204,809 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 85.10 |
| Mean single sequence MFE | -38.80 |
| Consensus MFE | -25.34 |
| Energy contribution | -27.65 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14204695 114 - 22224390 GCAUGUGGCAUGUGGCAUAUGGCACAAGCUGCUGCUUAAUGCGCCAGCCGGCAGCCAACUAAAUGCGACAUGUACGAGGAGUAAUGAGAGCGAACCAUCUGCAGAGAAGGUUCA ((((((.((((.((((.....(((.((((....))))..)))(((....))).)))).....)))).))))))..................(((((.(((....))).))))). ( -40.80) >DroSec_CAF1 116013 112 - 1 GCAUGUGGCAUGUGGCAUAUGGCACAAGCUGCUGCUUAAUGCACCAGCCGCUAGCCAACUAAAUGCGACAUGUACGAGUAGUAAUGAGAGCGAACCAUCUGCAG--AAGGUUCG ((((((.((((.((((...((((....((((.(((.....))).)))).)))))))).....)))).)))))).................((((((.((....)--).)))))) ( -40.10) >DroSim_CAF1 92039 112 - 1 GCAUGUGGCAUGUGGCAUAUGGCACAAGCUGCUGCUUAAUGCACCAGCCGCCAGCCAACUAAAUGCGACAUGUACGAGUAGUAAUGAGAGCGAACCAUCUGCAG--AAGGUUCG ((((((.((((.((((...((((....((((.(((.....))).)))).)))))))).....)))).)))))).................((((((.((....)--).)))))) ( -42.40) >DroYak_CAF1 105976 96 - 1 GCAUAUGGCAUACGGC-------ACUGG---CUGCUUAAUGCACCAGCCGCCAGUCAGCUAAAUGCGACAUGUACGAGGAGUAAUG------AACUAUCUGCAG--ACGGUCCA ((((.((((..(((((-------.((((---.(((.....)))))))..))).))..)))).))))(((.((((.((..(((....------.))).)))))).--...))).. ( -31.90) >consensus GCAUGUGGCAUGUGGCAUAUGGCACAAGCUGCUGCUUAAUGCACCAGCCGCCAGCCAACUAAAUGCGACAUGUACGAGGAGUAAUGAGAGCGAACCAUCUGCAG__AAGGUUCA ((((((.((((.((((...((((....((((.(((.....))).)))).)))))))).....)))).))))))..................(((((............))))). (-25.34 = -27.65 + 2.31)


| Location | 14,204,729 – 14,204,838 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 77.54 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -19.38 |
| Energy contribution | -22.76 |
| Covariance contribution | 3.38 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14204729 109 - 22224390 GCGUUUUUAAACGCGUCAUUUGCCAAAUUGCAUGUGGCAUGUGGCAUAUGGCACAAGCUGCUGCUUAAUGCGCCAGCCGGCAGCCAACUAAAUGCGACAUGUACGAGGA (((((....)))))........((....(((((((.((((.((((.....(((.((((....))))..)))(((....))).)))).....)))).)))))))...)). ( -38.80) >DroSec_CAF1 116045 109 - 1 GCGUUUUUAAACGCGUCAUUUGCCAAAUUGCAUGUGGCAUGUGGCAUAUGGCACAAGCUGCUGCUUAAUGCACCAGCCGCUAGCCAACUAAAUGCGACAUGUACGAGUA (((((....)))))...(((((......(((((((.((((.((((...((((....((((.(((.....))).)))).)))))))).....)))).)))))))))))). ( -37.70) >DroSim_CAF1 92071 109 - 1 GCGUUUUUAAACGCGUCAUUUGCCAAAUUGCAUGUGGCAUGUGGCAUAUGGCACAAGCUGCUGCUUAAUGCACCAGCCGCCAGCCAACUAAAUGCGACAUGUACGAGUA (((((....)))))...(((((......(((((((.((((.((((...((((....((((.(((.....))).)))).)))))))).....)))).)))))))))))). ( -40.00) >DroEre_CAF1 103710 87 - 1 GCGUUUUUAAAGCCCUCAUUUGCCAAAUUGCAUGUGCCAUGUGGCAU-----ACUAG---CUGCUUCAUGCACC--------------GAAAUGCGGCAUGUACGAGGA .............((((...(((......))).((((.((((.((((-----....(---.(((.....))).)--------------...)))).)))))))))))). ( -25.30) >DroYak_CAF1 106002 85 - 1 GCGUUUUUAAACGC--------------GGCAUAUGGCAUACGGC-------ACUGG---CUGCUUAAUGCACCAGCCGCCAGUCAGCUAAAUGCGACAUGUACGAGGA (((((....)))))--------------.((((.((((..(((((-------.((((---.(((.....)))))))..))).))..)))).)))).............. ( -29.70) >consensus GCGUUUUUAAACGCGUCAUUUGCCAAAUUGCAUGUGGCAUGUGGCAUAUGGCACAAGCUGCUGCUUAAUGCACCAGCCGCCAGCCAACUAAAUGCGACAUGUACGAGGA (((((....)))))..............(((((((.((((.(((....((((....((((.(((.....))).)))).....)))).))).)))).)))))))...... (-19.38 = -22.76 + 3.38)



| Location | 14,204,769 – 14,204,878 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 82.35 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -16.76 |
| Energy contribution | -19.56 |
| Covariance contribution | 2.80 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14204769 109 + 22224390 AUUAAGCAGCAGCUUGUGCCAUAUGCCACAUGCCACAUGCAAUUUGGCAAAUGACGCGUUUAAAAACGCGACAGUGGCAAUAAACACGCGCAAAACGUUUGAAAAUGAG .((((((....((.((((.....((((((.(((((.........)))))..((.((((((....)))))).)))))))).....)))).)).....))))))....... ( -32.40) >DroSec_CAF1 116085 109 + 1 AUUAAGCAGCAGCUUGUGCCAUAUGCCACAUGCCACAUGCAAUUUGGCAAAUGACGCGUUUAAAAACGCGACAGUGGCAAUAAACACGCGCGAAACGUUUGAAAAUGAG .((((((....((.((((.....((((((.(((((.........)))))..((.((((((....)))))).)))))))).....)))).)).....))))))....... ( -33.50) >DroSim_CAF1 92111 109 + 1 AUUAAGCAGCAGCUUGUGCCAUAUGCCACAUGCCACAUGCAAUUUGGCAAAUGACGCGUUUAAAAACGCGACAGUGGCAAUAAACACGCGCGGAACGUUUGAAAAUGAG .((((((..(.((.((((.....((((((.(((((.........)))))..((.((((((....)))))).)))))))).....)))).)).)...))))))....... ( -33.70) >DroEre_CAF1 103736 101 + 1 AUGAAGCAG---CUAGU-----AUGCCACAUGGCACAUGCAAUUUGGCAAAUGAGGGCUUUAAAAACGCGACAUUGGCAAUAAACACGCGCGAAGCGUUUGGAAAUGAG .((((((..---((.((-----.(((((.((.((....)).)).))))).)).)).)))))).((((((....(((((.........)).))).))))))......... ( -25.50) >DroYak_CAF1 106042 85 + 1 AUUAAGCAG---CCAGU-------GCCGUAUGCCAUAUGCC--------------GCGUUUAAAAACGCGACAGCGGCAAUAAACACGCGCAAAACGUUUGAAAACGCG .....((..---....(-------(((((..((.....))(--------------(((((....))))))...)))))).....((.(((.....))).)).....)). ( -22.00) >consensus AUUAAGCAGCAGCUUGUGCCAUAUGCCACAUGCCACAUGCAAUUUGGCAAAUGACGCGUUUAAAAACGCGACAGUGGCAAUAAACACGCGCGAAACGUUUGAAAAUGAG .((((((....((.((((.....((((((.(((((.........)))))......(((((....)))))....)))))).....)))).)).....))))))....... (-16.76 = -19.56 + 2.80)



| Location | 14,204,769 – 14,204,878 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 82.35 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -17.26 |
| Energy contribution | -20.18 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14204769 109 - 22224390 CUCAUUUUCAAACGUUUUGCGCGUGUUUAUUGCCACUGUCGCGUUUUUAAACGCGUCAUUUGCCAAAUUGCAUGUGGCAUGUGGCAUAUGGCACAAGCUGCUGCUUAAU ......................(((((...((((((....(((((....)))))(((((.(((......))).)))))..))))))...)))))((((....))))... ( -34.60) >DroSec_CAF1 116085 109 - 1 CUCAUUUUCAAACGUUUCGCGCGUGUUUAUUGCCACUGUCGCGUUUUUAAACGCGUCAUUUGCCAAAUUGCAUGUGGCAUGUGGCAUAUGGCACAAGCUGCUGCUUAAU ......................(((((...((((((....(((((....)))))(((((.(((......))).)))))..))))))...)))))((((....))))... ( -34.60) >DroSim_CAF1 92111 109 - 1 CUCAUUUUCAAACGUUCCGCGCGUGUUUAUUGCCACUGUCGCGUUUUUAAACGCGUCAUUUGCCAAAUUGCAUGUGGCAUGUGGCAUAUGGCACAAGCUGCUGCUUAAU ......................(((((...((((((....(((((....)))))(((((.(((......))).)))))..))))))...)))))((((....))))... ( -34.60) >DroEre_CAF1 103736 101 - 1 CUCAUUUCCAAACGCUUCGCGCGUGUUUAUUGCCAAUGUCGCGUUUUUAAAGCCCUCAUUUGCCAAAUUGCAUGUGCCAUGUGGCAU-----ACUAG---CUGCUUCAU ..................((((((((.....((.((((..((.........))...)))).))......))))))))...(..((..-----....)---)..)..... ( -21.80) >DroYak_CAF1 106042 85 - 1 CGCGUUUUCAAACGUUUUGCGCGUGUUUAUUGCCGCUGUCGCGUUUUUAAACGC--------------GGCAUAUGGCAUACGGC-------ACUGG---CUGCUUAAU .(((((....)))))...(((((((((...(((((.(((((((((....)))))--------------))))..)))))...)))-------))..)---).))..... ( -29.30) >consensus CUCAUUUUCAAACGUUUCGCGCGUGUUUAUUGCCACUGUCGCGUUUUUAAACGCGUCAUUUGCCAAAUUGCAUGUGGCAUGUGGCAUAUGGCACAAGCUGCUGCUUAAU .............((...((..(((((...(((((((((((((((....)))))......(((......)))...)))).))))))...)))))..))....))..... (-17.26 = -20.18 + 2.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:33 2006