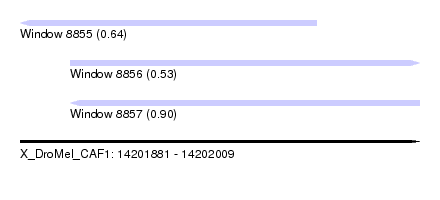
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,201,881 – 14,202,009 |
| Length | 128 |
| Max. P | 0.898215 |

| Location | 14,201,881 – 14,201,976 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 91.37 |
| Mean single sequence MFE | -25.65 |
| Consensus MFE | -22.33 |
| Energy contribution | -23.08 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14201881 95 - 22224390 CAUUCGCAU-UCGCACAUCGCUCAUACGCCGUGUGGUACGCGCUGGCAAUCAACCUGCAAGUUGCCAGCCUAAUGUUUAAUUAAACAGCUCACCGU .....((..-..))((((.((......)).))))(((..(((((((((((..........)))))))))....(((((....)))))))..))).. ( -26.70) >DroSec_CAF1 111755 95 - 1 AAAUCGCAU-UCGCACAUCGCUCAUACGCCGUGUGGUGCGCGGUGGCAAUCGAGCUGCAAGUUGCCAGCCUAAUGUUUAAUUAAACAGCUCACCGU .....((((-.(((((...((......)).)))))))))((((((((......(((((.....).))))....(((((....))))))).)))))) ( -30.00) >DroEre_CAF1 100752 90 - 1 -----GCAU-UUGCACAUCGCUCAUACGCCGUGUGGUACGCGCUGGCAAUCAACCUGCAAGUUAGCAGCCUAAUGUUUAAUUAAACAGCUCACCGU -----....-...(((((.((......)).)))))....(((.((((.......((((......)))).....(((((....))))))).)).))) ( -19.50) >DroYak_CAF1 103045 91 - 1 -----GCACAUCGCACAUCGCUCAUACGCCGUGUGGUACGCGCUGGCAAUCAACCUGCAAGUUGCCAGCCUAAUGUUUAAUUAAACAGCUCACCGU -----((.....))((((.((......)).))))(((..(((((((((((..........)))))))))....(((((....)))))))..))).. ( -26.40) >consensus _____GCAU_UCGCACAUCGCUCAUACGCCGUGUGGUACGCGCUGGCAAUCAACCUGCAAGUUGCCAGCCUAAUGUUUAAUUAAACAGCUCACCGU .....((.....))((((.((......)).))))(((..(((((((((((..........)))))))))....(((((....)))))))..))).. (-22.33 = -23.08 + 0.75)

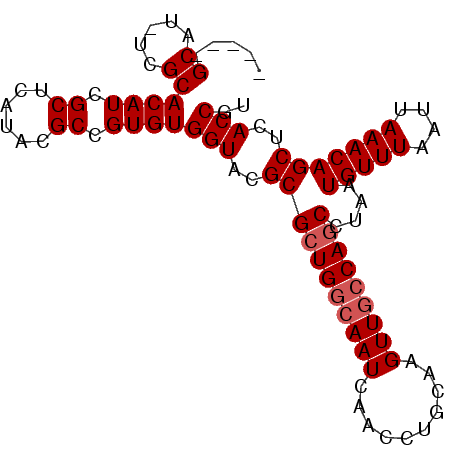
| Location | 14,201,897 – 14,202,009 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 91.03 |
| Mean single sequence MFE | -38.78 |
| Consensus MFE | -29.90 |
| Energy contribution | -30.15 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14201897 112 + 22224390 UUAAACAUUAGGCUGGCAACUUGCAGGUUGAUUGCCAGCGCGUACCACACGGCGUAUGAGCGAUGUGCGA-AUGCGAAUGUGAAUGCGAAUGCCAGUAGUUGCGGUUGCCAGC ...........((((((((((.((((.(.....((...(((((..((((...(((((..((.....))..-)))))..)))).)))))...))....).)))))))))))))) ( -41.30) >DroSec_CAF1 111771 112 + 1 UUAAACAUUAGGCUGGCAACUUGCAGCUCGAUUGCCACCGCGCACCACACGGCGUAUGAGCGAUGUGCGA-AUGCGAUUUCGAAUGCGAAUGCCAGUAGUUGCGGUUGCCAGC ...........((((((((((.((((((..((((.((...((((.((((..((......))..))))(((-(......))))..))))..)).)))))))))))))))))))) ( -42.10) >DroEre_CAF1 100768 106 + 1 UUAAACAUUAGGCUGCUAACUUGCAGGUUGAUUGCCAGCGCGUACCACACGGCGUAUGAGCGAUGUGCAA-AUGC------GAAUGUGAAUGCCAGUAGUUGCGGCUGCCAGC ..........((((((.((((.((.(((...(..(...(((((..((((..((......))..))))...-))))------)...)..)..))).))))))))))))...... ( -31.50) >DroYak_CAF1 103061 107 + 1 UUAAACAUUAGGCUGGCAACUUGCAGGUUGAUUGCCAGCGCGUACCACACGGCGUAUGAGCGAUGUGCGAUGUGC------GAAUGCGAAUGCCAGUAGUUGCGGUUGCCAGC ...........((((((((((.((((((...((((..((((((..((((..((......))..))))..))))))------....))))..)))......))))))))))))) ( -40.20) >consensus UUAAACAUUAGGCUGGCAACUUGCAGGUUGAUUGCCAGCGCGUACCACACGGCGUAUGAGCGAUGUGCGA_AUGC______GAAUGCGAAUGCCAGUAGUUGCGGUUGCCAGC ...........((((((((((.((((....(((((((..((((........)))).)).)))))(.(((...((((........))))..)))).....)))))))))))))) (-29.90 = -30.15 + 0.25)



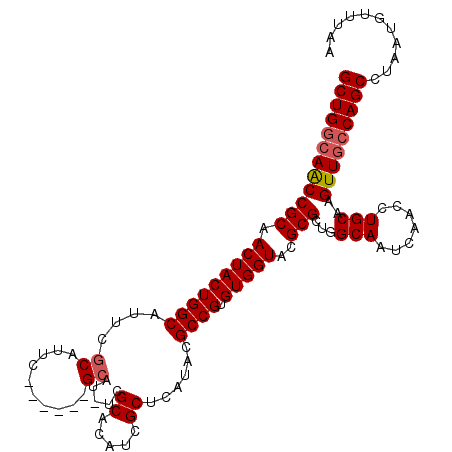
| Location | 14,201,897 – 14,202,009 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 91.03 |
| Mean single sequence MFE | -35.15 |
| Consensus MFE | -30.51 |
| Energy contribution | -31.08 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14201897 112 - 22224390 GCUGGCAACCGCAACUACUGGCAUUCGCAUUCACAUUCGCAU-UCGCACAUCGCUCAUACGCCGUGUGGUACGCGCUGGCAAUCAACCUGCAAGUUGCCAGCCUAAUGUUUAA (((((((((.((.(((((((((....((..........))..-..((.....))......)))).)))))..))((.((.......)).))..)))))))))........... ( -35.10) >DroSec_CAF1 111771 112 - 1 GCUGGCAACCGCAACUACUGGCAUUCGCAUUCGAAAUCGCAU-UCGCACAUCGCUCAUACGCCGUGUGGUGCGCGGUGGCAAUCGAGCUGCAAGUUGCCAGCCUAAUGUUUAA ((((((((((((.(((((((((.((((....))))...((..-..)).............)))).)))))..)))(..((......))..)..)))))))))........... ( -42.10) >DroEre_CAF1 100768 106 - 1 GCUGGCAGCCGCAACUACUGGCAUUCACAUUC------GCAU-UUGCACAUCGCUCAUACGCCGUGUGGUACGCGCUGGCAAUCAACCUGCAAGUUAGCAGCCUAAUGUUUAA ((..((.((((.......))))..........------((..-...(((((.((......)).)))))....))))..)).......((((......))))............ ( -27.40) >DroYak_CAF1 103061 107 - 1 GCUGGCAACCGCAACUACUGGCAUUCGCAUUC------GCACAUCGCACAUCGCUCAUACGCCGUGUGGUACGCGCUGGCAAUCAACCUGCAAGUUGCCAGCCUAAUGUUUAA (((((((((.((.(((((((((....((....------)).....((.....))......)))).)))))..))((.((.......)).))..)))))))))........... ( -36.00) >consensus GCUGGCAACCGCAACUACUGGCAUUCGCAUUC______GCAU_UCGCACAUCGCUCAUACGCCGUGUGGUACGCGCUGGCAAUCAACCUGCAAGUUGCCAGCCUAAUGUUUAA ((((((((((((.(((((((((....((..........)).....((.....))......)))).)))))..)))...(((.......)))..)))))))))........... (-30.51 = -31.08 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:28 2006