
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,201,479 – 14,201,583 |
| Length | 104 |
| Max. P | 0.992989 |

| Location | 14,201,479 – 14,201,583 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.78 |
| Mean single sequence MFE | -44.28 |
| Consensus MFE | -23.98 |
| Energy contribution | -26.33 |
| Covariance contribution | 2.35 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
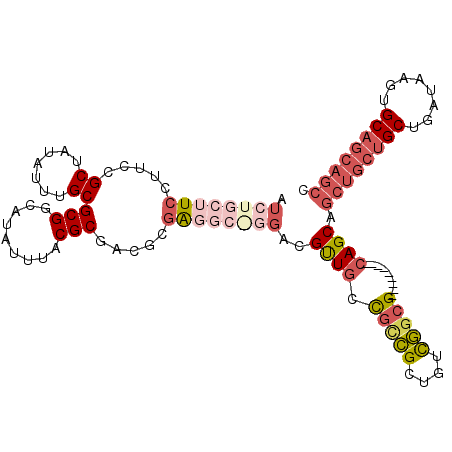
>X_DroMel_CAF1 14201479 104 + 22224390 AUCUGCUUCCUUCCGCUAUAUUUGCGCGGCAUAUUUACGCGACGCGAGGCGGGACGCUGCUGCCGCUGUCGGCG---------CAGCAGCUGCUGCUGAUAAGUGCAGCAGCC .(((((((((.((.((.......))(((.........))))).).))))))))..(((((.((((....)))))---------)))).((((((((........)))))))). ( -48.30) >DroPse_CAF1 124104 110 + 1 AUCUGCUUCCUUCCGCCAUAUUUGCGCGGCAUAUUUACGCGUUGCUGC---CGCCGUUGCCGUUGCCGUCAGCGUAGCAGCGGCAGCAGCUGCUGCUGAUAAGUGCAGCAGCC ....((......((((((....)).)))).........))..((((((---(((.(((((.((((....))))))))).)))))))))((((((((........)))))))). ( -50.46) >DroSec_CAF1 111391 91 + 1 AUCUGCUUCCUUCCGCUAUAUUUGCG-------------CGAAGCGAGGCGGGACGCUGCUGCCGCUGUCGGCG---------CAGCAGCUGCUGCUGAUAAGUGCAGCAGCC .(((((((((((((((.......)))-------------.)))).))))))))..(((((.((((....)))))---------)))).((((((((........)))))))). ( -52.00) >DroEre_CAF1 100363 104 + 1 AUCUGCUUCCUUCCGCUAUAUUUGCGCGGCAUAUUUACGCGACGCGGGGCAGGACGUUGCCGCCGCUGUCGGCG---------CAGCAGCUGCUGCUGAUAAGUGCAGCAGCC .(((((((((.((.((.......))(((.........))))).).))))))))..(((((.((((....)))))---------)))).((((((((........)))))))). ( -45.60) >DroWil_CAF1 84973 82 + 1 AUCUGCUUCCUUCCGCCAUAUUUGCGCGGCAUAUUUACGCGUCGUGG-------------CUUCGCUGUUGGCG---------U---------CGCUGAUAAGUGCAGGCAGC ..((((((......(((((....(((((.........))))).))))-------------)..((((((..((.---------.---------.))..)).)))).)))))). ( -26.10) >DroYak_CAF1 102638 104 + 1 AUCUGCUUCCUUCCGCUAUAUUUGCGCGGCAUAUUUACGCGACGCGAGGCAGGACGUUGCUGCCGCUGUCGACG---------CAGCAGCUGCUGCUGAUAAGUGCAGCAGCC .(((((((((.((.((.......))(((.........))))).).))))))))..((((((((.(((((((..(---------((((....))))))))).))))))))))). ( -43.20) >consensus AUCUGCUUCCUUCCGCUAUAUUUGCGCGGCAUAUUUACGCGACGCGAGGC_GGACGUUGCCGCCGCUGUCGGCG_________CAGCAGCUGCUGCUGAUAAGUGCAGCAGCC .((((((((.....((.......))(((.........))).....))))))))..((((.(((((....))))).........)))).((((((((........)))))))). (-23.98 = -26.33 + 2.35)


| Location | 14,201,479 – 14,201,583 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.78 |
| Mean single sequence MFE | -43.48 |
| Consensus MFE | -21.92 |
| Energy contribution | -24.22 |
| Covariance contribution | 2.29 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.50 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.992989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
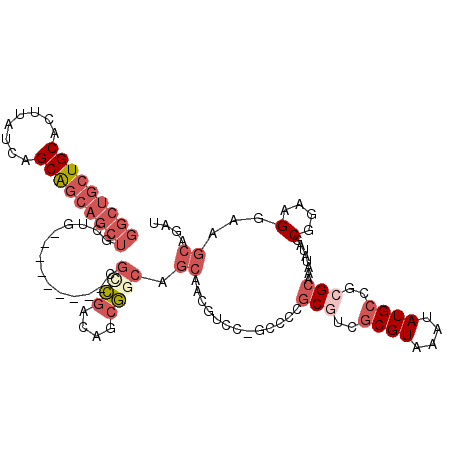
>X_DroMel_CAF1 14201479 104 - 22224390 GGCUGCUGCACUUAUCAGCAGCAGCUGCUG---------CGCCGACAGCGGCAGCAGCGUCCCGCCUCGCGUCGCGUAAAUAUGCCGCGCAAAUAUAGCGGAAGGAAGCAGAU (((((((((........)))))))))((((---------(((((....)))).))))).....((.((((((.((((....)))).))))........(....))).)).... ( -48.10) >DroPse_CAF1 124104 110 - 1 GGCUGCUGCACUUAUCAGCAGCAGCUGCUGCCGCUGCUACGCUGACGGCAACGGCAACGGCG---GCAGCAACGCGUAAAUAUGCCGCGCAAAUAUGGCGGAAGGAAGCAGAU (((((((((........)))))))))((((((((((((........)))).((....)))))---)))))...((.........((((.((....))))))......)).... ( -49.96) >DroSec_CAF1 111391 91 - 1 GGCUGCUGCACUUAUCAGCAGCAGCUGCUG---------CGCCGACAGCGGCAGCAGCGUCCCGCCUCGCUUCG-------------CGCAAAUAUAGCGGAAGGAAGCAGAU (((((((((........)))))))))((((---------(((((....)))).))))).....((.((.((((.-------------(((.......))))))))).)).... ( -45.40) >DroEre_CAF1 100363 104 - 1 GGCUGCUGCACUUAUCAGCAGCAGCUGCUG---------CGCCGACAGCGGCGGCAACGUCCUGCCCCGCGUCGCGUAAAUAUGCCGCGCAAAUAUAGCGGAAGGAAGCAGAU (((((((((........))))))))).(((---------(((((((.((((.((((......)))))))))))).)).......((((.........))))......)))).. ( -49.80) >DroWil_CAF1 84973 82 - 1 GCUGCCUGCACUUAUCAGCG---------A---------CGCCAACAGCGAAG-------------CCACGACGCGUAAAUAUGCCGCGCAAAUAUGGCGGAAGGAAGCAGAU (((.(((((........)).---------.---------(((((...(((...-------------...)).)((((.........)))).....)))))..))).))).... ( -21.20) >DroYak_CAF1 102638 104 - 1 GGCUGCUGCACUUAUCAGCAGCAGCUGCUG---------CGUCGACAGCGGCAGCAACGUCCUGCCUCGCGUCGCGUAAAUAUGCCGCGCAAAUAUAGCGGAAGGAAGCAGAU .(((((((.......))))))).(((((((---------(.......))))))))......((((.((((((.((((....)))).))))........(....))).)))).. ( -46.40) >consensus GGCUGCUGCACUUAUCAGCAGCAGCUGCUG_________CGCCGACAGCGGCAGCAACGUCC_GCCCCGCGUCGCGUAAAUAUGCCGCGCAAAUAUAGCGGAAGGAAGCAGAU (((((((((........)))))))))..............((((....)))).((.............(((..((((....))))..)))........(....)...)).... (-21.92 = -24.22 + 2.29)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:25 2006