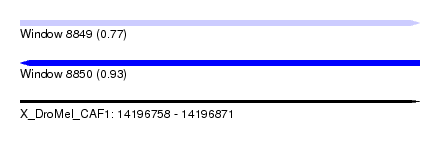
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,196,758 – 14,196,871 |
| Length | 113 |
| Max. P | 0.934131 |

| Location | 14,196,758 – 14,196,871 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 95.40 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -26.34 |
| Energy contribution | -26.46 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14196758 113 + 22224390 GCGUGUCCUGGGCCAUAGUUCAAUUUUAAGCAUCGGAGACAGCAGAAGCAUUUUUGCUUCGCAUUCUUUUUGGUUGUUGAUGUUGACCAUGGCAUGUUUGACAAUUUCCACUU ...((((....(((((.((.((((....(((((((((((..((.((((((....))))))))....))))))).))))...)))))).)))))......)))).......... ( -30.60) >DroSec_CAF1 106881 113 + 1 GCGUGUCCUGGGCUAUAGUUCAAUUUUAAGCAUCGGAGACAGCAGAAGCAUUUUUGCUUCACAUUCUUUUUGGUUGUUGAUGUUGACCAUGGCAUGUUUGGCAAUUUCCACUU (((((((.(((((....)).........((((((...((((((.((((((....)))))).((.......))))))))))))))..))).))))))).(((......)))... ( -30.50) >DroSim_CAF1 84552 113 + 1 GCGUGUCCUGGGCCAUAGUUCAAUUUUAAGCAUCGGAGACAGCAGAAGCAUUUUUGCUUCGCAUUCUUUUUGGUUGUUGAUGUUGACCAUGGCAUGUUUGACAAUUUCCACUU ...((((....(((((.((.((((....(((((((((((..((.((((((....))))))))....))))))).))))...)))))).)))))......)))).......... ( -30.60) >DroEre_CAF1 95882 110 + 1 ---UGUCCUGGGCCAUAGUUCAAUUUUAAGCAUCGGAGACUGCAGAAGCAUUUCUGCCCCGCAUUCUUUUUGCUUGUUGAUGUUGACCAUGGCAUGUUUGACAAUUUCCACUU ---((((....(((((.((.((((..((((((..(((((.(((.(..(((....)))..)))).))))).)))))).....)))))).)))))......)))).......... ( -29.10) >DroYak_CAF1 97980 113 + 1 GCGUGUCCAGGGCCAUAGUUCAAUUUUAAGCAUCGGAGACAGCAGAAGCAUUUUUGUUUCGCAUUCUUUUUGGUUGUUGAUGUUGACCAUGGCAUGUUUGACAAUUUCCACUU ...((((((..(((((.((.((((....(((((((((((..((.((((((....))))))))....))))))).))))...)))))).))))).))...)))).......... ( -28.30) >consensus GCGUGUCCUGGGCCAUAGUUCAAUUUUAAGCAUCGGAGACAGCAGAAGCAUUUUUGCUUCGCAUUCUUUUUGGUUGUUGAUGUUGACCAUGGCAUGUUUGACAAUUUCCACUU ...((((....(((((.((.((((....(((((((((((.....((((((....))))))......))))))).))))...)))))).)))))......)))).......... (-26.34 = -26.46 + 0.12)


| Location | 14,196,758 – 14,196,871 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 95.40 |
| Mean single sequence MFE | -25.34 |
| Consensus MFE | -22.46 |
| Energy contribution | -22.58 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14196758 113 - 22224390 AAGUGGAAAUUGUCAAACAUGCCAUGGUCAACAUCAACAACCAAAAAGAAUGCGAAGCAAAAAUGCUUCUGCUGUCUCCGAUGCUUAAAAUUGAACUAUGGCCCAGGACACGC ..........((((......((((((((..................(((..((((((((....)))))).))..))).((((.......)))).))))))))....))))... ( -25.90) >DroSec_CAF1 106881 113 - 1 AAGUGGAAAUUGCCAAACAUGCCAUGGUCAACAUCAACAACCAAAAAGAAUGUGAAGCAAAAAUGCUUCUGCUGUCUCCGAUGCUUAAAAUUGAACUAUAGCCCAGGACACGC ..(((((..((((((.........))).)))..)).....((...........((((((....)))))).(((((...((((.......))))....)))))...)).))).. ( -20.50) >DroSim_CAF1 84552 113 - 1 AAGUGGAAAUUGUCAAACAUGCCAUGGUCAACAUCAACAACCAAAAAGAAUGCGAAGCAAAAAUGCUUCUGCUGUCUCCGAUGCUUAAAAUUGAACUAUGGCCCAGGACACGC ..........((((......((((((((..................(((..((((((((....)))))).))..))).((((.......)))).))))))))....))))... ( -25.90) >DroEre_CAF1 95882 110 - 1 AAGUGGAAAUUGUCAAACAUGCCAUGGUCAACAUCAACAAGCAAAAAGAAUGCGGGGCAGAAAUGCUUCUGCAGUCUCCGAUGCUUAAAAUUGAACUAUGGCCCAGGACA--- ..........((((......(((((((((((.......(((((...(((.(((((((((....))).)))))).)))....)))))....))).))))))))....))))--- ( -34.30) >DroYak_CAF1 97980 113 - 1 AAGUGGAAAUUGUCAAACAUGCCAUGGUCAACAUCAACAACCAAAAAGAAUGCGAAACAAAAAUGCUUCUGCUGUCUCCGAUGCUUAAAAUUGAACUAUGGCCCUGGACACGC ..........((((......(((((((((((((((......((...((((.(((.........)))))))..)).....)))).......))).))))))))....))))... ( -20.11) >consensus AAGUGGAAAUUGUCAAACAUGCCAUGGUCAACAUCAACAACCAAAAAGAAUGCGAAGCAAAAAUGCUUCUGCUGUCUCCGAUGCUUAAAAUUGAACUAUGGCCCAGGACACGC ..........((((......((((((((..................(((..((((((((....)))))).))..))).((((.......)))).))))))))....))))... (-22.46 = -22.58 + 0.12)

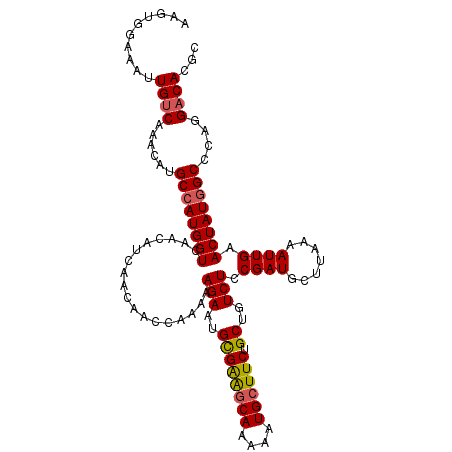
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:22 2006