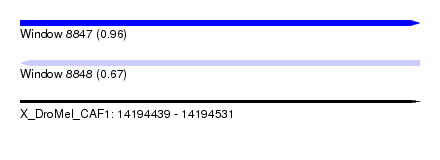
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,194,439 – 14,194,531 |
| Length | 92 |
| Max. P | 0.963614 |

| Location | 14,194,439 – 14,194,531 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 90.59 |
| Mean single sequence MFE | -31.74 |
| Consensus MFE | -24.46 |
| Energy contribution | -24.54 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
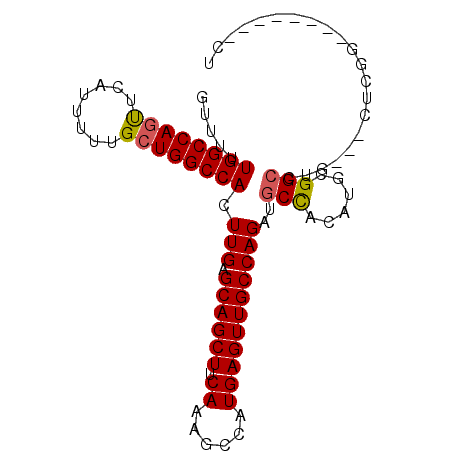
>X_DroMel_CAF1 14194439 92 + 22224390 GUUUUUGGCCAGUUCAUUUUUGCUGGCCACUUGAGCAGCUUCAAAGCCAUGAGUUGCCAGAUGCCACAUGCGGCCUG---CUCGGCUGUUCGGCU .....((((((((........)))))))).(((.((((((.((......)))))))))))..(((....((((((..---...))))))..))). ( -36.50) >DroSec_CAF1 104701 84 + 1 GUUUUUGGCCAGUUCAUUUUUGCUGGCCACUUGAGCAGCUUCAACGCCAUGAGUUGCCAGAUGCCACAUGCGGCCUG---CUCGG--------CU .....((((((((........)))))))).((((((((...((((.......))))......(((......))))))---)))))--------.. ( -30.00) >DroSim_CAF1 82281 84 + 1 GUUUUUGGCCAGUUCAUUUUUGCUGGCCACUUGAGCAGCUUCAAAGCCAUGAGUUGCCAGAUGCCACAUGCGGCCUG---CUCGG--------CU ((((.((((((((........))))))))...))))........((((..((((.(((..(((...)))..)))..)---)))))--------)) ( -29.90) >DroEre_CAF1 93783 86 + 1 GUUUUUGGCCAGUUCAUUUUUGCUGGCCACUUGAGCAGCUUCAAAGCCAUGAGUUGCCAGCUGCUACAUGCGG-CUGCUGUUGGG--------CU .....((((((((........))))))))((..(((((((.((......))))))))(((((((.....))))-)))...)..))--------.. ( -34.90) >DroYak_CAF1 95720 87 + 1 GUUUUUGGCCAACUCAUUUUUGCUGGCCACUUGAGCAGCUUCAAAGCCAUGAGUUGCCAGAUGCCACAUGCGGCCUGCUGUUCGG--------CU ...((((((.(((((((...((((.(.....).))))(((....))).))))))))))))).(((....((((....))))..))--------). ( -27.40) >consensus GUUUUUGGCCAGUUCAUUUUUGCUGGCCACUUGAGCAGCUUCAAAGCCAUGAGUUGCCAGAUGCCACAUGCGGCCUG___CUCGG________CU .....((((((((........)))))))).(((.((((((.((......)))))))))))..(((......)))..................... (-24.46 = -24.54 + 0.08)


| Location | 14,194,439 – 14,194,531 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 90.59 |
| Mean single sequence MFE | -29.54 |
| Consensus MFE | -20.52 |
| Energy contribution | -20.72 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
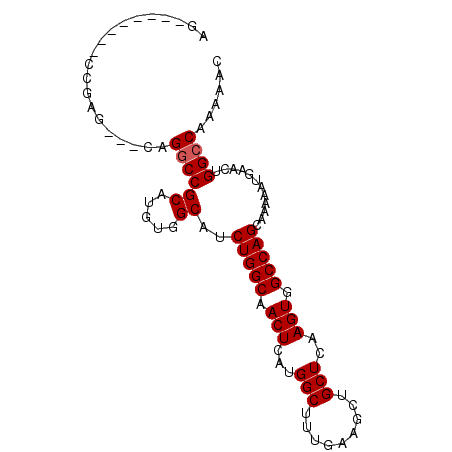
>X_DroMel_CAF1 14194439 92 - 22224390 AGCCGAACAGCCGAG---CAGGCCGCAUGUGGCAUCUGGCAACUCAUGGCUUUGAAGCUGCUCAAGUGGCCAGCAAAAAUGAACUGGCCAAAAAC ............(((---(((((((....)))).((.(((........)))..))..))))))...(((((((..........)))))))..... ( -30.40) >DroSec_CAF1 104701 84 - 1 AG--------CCGAG---CAGGCCGCAUGUGGCAUCUGGCAACUCAUGGCGUUGAAGCUGCUCAAGUGGCCAGCAAAAAUGAACUGGCCAAAAAC ..--------..(((---(((((((....))))..((..((((.......)))).))))))))...(((((((..........)))))))..... ( -30.20) >DroSim_CAF1 82281 84 - 1 AG--------CCGAG---CAGGCCGCAUGUGGCAUCUGGCAACUCAUGGCUUUGAAGCUGCUCAAGUGGCCAGCAAAAAUGAACUGGCCAAAAAC ..--------..(((---(((((((....)))).((.(((........)))..))..))))))...(((((((..........)))))))..... ( -30.40) >DroEre_CAF1 93783 86 - 1 AG--------CCCAACAGCAG-CCGCAUGUAGCAGCUGGCAACUCAUGGCUUUGAAGCUGCUCAAGUGGCCAGCAAAAAUGAACUGGCCAAAAAC ..--------......(((((-(..((...(((....(....).....))).))..))))))....(((((((..........)))))))..... ( -26.80) >DroYak_CAF1 95720 87 - 1 AG--------CCGAACAGCAGGCCGCAUGUGGCAUCUGGCAACUCAUGGCUUUGAAGCUGCUCAAGUGGCCAGCAAAAAUGAGUUGGCCAAAAAC ..--------..((.((((..((((....)))).((.(((........)))..)).)))).))...((((((((........))))))))..... ( -29.90) >consensus AG________CCGAG___CAGGCCGCAUGUGGCAUCUGGCAACUCAUGGCUUUGAAGCUGCUCAAGUGGCCAGCAAAAAUGAACUGGCCAAAAAC ....................((((((.....))..(((((.(((...(((.........)))..))).)))))............))))...... (-20.52 = -20.72 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:20 2006