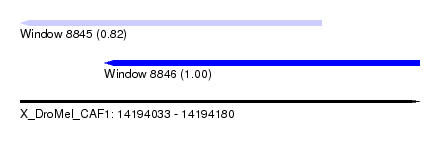
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,194,033 – 14,194,180 |
| Length | 147 |
| Max. P | 0.996887 |

| Location | 14,194,033 – 14,194,144 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 86.06 |
| Mean single sequence MFE | -28.14 |
| Consensus MFE | -23.32 |
| Energy contribution | -22.52 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
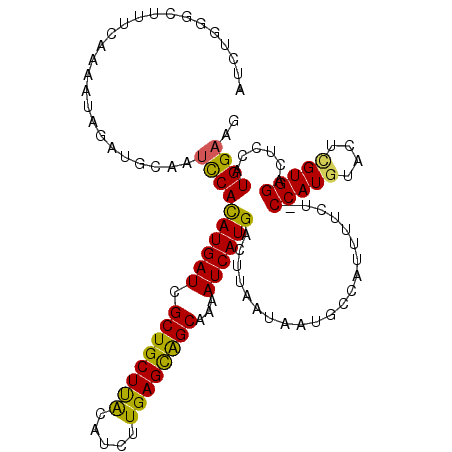
>X_DroMel_CAF1 14194033 111 - 22224390 AUCUGGGCUUUCACAAUAGAUGCAAUCCACAUGAUCGCUGCUUACAUUUUGAGCAGCAAAAUCAUGACUUAAUAAUGCCAUUUUCU-CCAUGUACUCGUGGA-UCCAUGGUAG ...(((((..((......)).))...)))((((((.((((((((.....))))))))...)))))).........((((((....(-(((((....))))))-...)))))). ( -32.20) >DroSec_CAF1 104290 113 - 1 AUCUGGGCUUUCAGAAGAGAUGCAAUCCAUAUGAUCGCUGCUUACAUCUUGAGCAGCAAAAUCAUGACGUAAUAAUGCCAUUUUCUCCCAUGUACUCGUGGACUCCAUGGAAG .......((((((((..(((((.......((((((.((((((((.....))))))))...))))))..(((....))))))))..))(((((....)))))......)))))) ( -28.60) >DroSim_CAF1 81865 113 - 1 AUCUGGGCUUUCAGUAGAGAUGCAAUCCACAUGAUCGCUGCUUACAUCUUGAGCAGCAAAAUCAUGACUUAAUAAUGCCAUUUUCUUCCAUGUACUGGUGGACUCCAUGGAAG ...(((((((((....)))).))......((((((.((((((((.....))))))))...))))))...........)))....((((((((....(.....)..)))))))) ( -31.70) >DroEre_CAF1 93432 103 - 1 -------UCUUUGAAGUAGAUGCCAUCCACAUGAUCGCUGCUCGCAUCUUGAGUGGCAAAAUCAUGACCUAAUAAUGCCUUUUUCU-CCAUGUACUUGUGGAC--CAUGGAAG -------..................((((((((((.((..((((.....))))..))...))))))...................(-((((......))))).--..)))).. ( -24.00) >DroYak_CAF1 95242 111 - 1 AUCUGGCUUUUUAAAAUAGAUGCAAUUCACAUGAUCGCUGCUUACAUCUUGAGUAGCAAAAUCAUGACUUAAUAAUGCCUUUUUCG-CCAUGUACUUGUGGAC-CCAUGGAAG ....(((...((((....((......)).((((((.((((((((.....))))))))...))))))..))))....))).......-(((((..(....)...-.)))))... ( -24.20) >consensus AUCUGGGCUUUCAAAAUAGAUGCAAUCCACAUGAUCGCUGCUUACAUCUUGAGCAGCAAAAUCAUGACUUAAUAAUGCCAUUUUCU_CCAUGUACUCGUGGACUCCAUGGAAG .........................((((((((((.((((((((.....))))))))...)))))).....................(((((....)))))......)))).. (-23.32 = -22.52 + -0.80)


| Location | 14,194,064 – 14,194,180 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.98 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -21.26 |
| Energy contribution | -21.74 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14194064 116 - 22224390 UC--AUA--AGUAGCUCAGAGAGAUAGAGAGUCCAGACUCAUCUGGGCUUUCACAAUAGAUGCAAUCCACAUGAUCGCUGCUUACAUUUUGAGCAGCAAAAUCAUGACUUAAUAAUGCCA ..--.((--(((.((((.....))..(((((((((((....))))))))))).........))......((((((.((((((((.....))))))))...)))))))))))......... ( -37.20) >DroSec_CAF1 104323 112 - 1 UC--AUA--AGCAGCUCAGUGA----GAGAGUACAGACUCAUCUGGGCUUUCAGAAGAGAUGCAAUCCAUAUGAUCGCUGCUUACAUCUUGAGCAGCAAAAUCAUGACGUAAUAAUGCCA ..--...--.(((.(((..(..----(((((..((((....))))..))))).)..))).)))......((((((.((((((((.....))))))))...)))))).............. ( -32.50) >DroSim_CAF1 81898 112 - 1 UC--AUA--AGUAACUCAGUGA----GAGAGUACAGACUCAUCUGGGCUUUCAGUAGAGAUGCAAUCCACAUGAUCGCUGCUUACAUCUUGAGCAGCAAAAUCAUGACUUAAUAAUGCCA ..--.((--(((..((((.(((----(((....((((....))))..)))))).).)))..........((((((.((((((((.....))))))))...)))))))))))......... ( -33.70) >DroEre_CAF1 93462 105 - 1 UUAAGUACGAGUAUCUCAGCGG------GCAUACGCUA---------UCUUUGAAGUAGAUGCCAUCCACAUGAUCGCUGCUCGCAUCUUGAGUGGCAAAAUCAUGACCUAAUAAUGCCU ........(((...))).((((------((((..(((.---------.......)))..))))).....((((((.((..((((.....))))..))...)))))).........))).. ( -27.30) >DroYak_CAF1 95273 112 - 1 UUUAAUA--AGUAGCUCAGCGA------GAAUACAGAUUCAUCUGGCUUUUUAAAAUAGAUGCAAUUCACAUGAUCGCUGCUUACAUCUUGAGUAGCAAAAUCAUGACUUAAUAAUGCCU .(((.((--(((..........------((((.((((....))))((.(((......))).)).)))).((((((.((((((((.....))))))))...))))))))))).)))..... ( -24.90) >consensus UC__AUA__AGUAGCUCAGCGA____GAGAGUACAGACUCAUCUGGGCUUUCAAAAUAGAUGCAAUCCACAUGAUCGCUGCUUACAUCUUGAGCAGCAAAAUCAUGACUUAAUAAUGCCA ..........(((.............((((((.(((......))).)))))).................((((((.((((((((.....))))))))...)))))).........))).. (-21.26 = -21.74 + 0.48)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:18 2006