
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,185,650 – 14,185,751 |
| Length | 101 |
| Max. P | 0.974429 |

| Location | 14,185,650 – 14,185,751 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 88.33 |
| Mean single sequence MFE | -38.95 |
| Consensus MFE | -31.70 |
| Energy contribution | -32.20 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
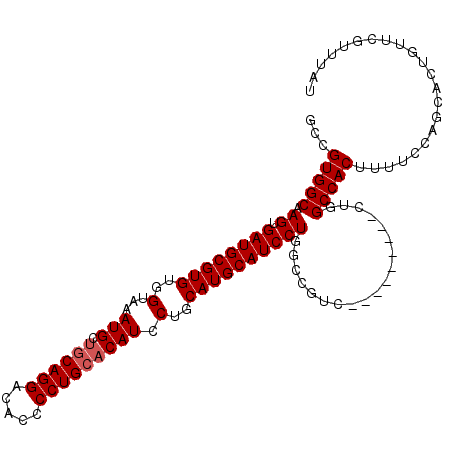
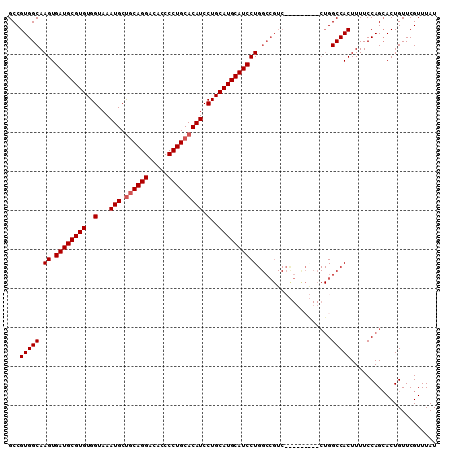
>X_DroMel_CAF1 14185650 101 + 22224390 GCCGUGGCAAGUGAUGCGUGUGGUAAAUGCCGCAGGACACCCCUGCACAUCCUGCAUGCAUCCUGGCCGUC---------CUGGCCACUUUUCCAGCACUGUUCGUUUAU .....(((.((.((((((((((((....)))(((((.....))))).......))))))))))).)))((.---------((((........)))).))........... ( -37.30) >DroSec_CAF1 96053 110 + 1 GCCGUGGCAAGUGAUGCGUGUGGUAAAUGCUGCAGGACACCCCUGCACAUCCUGCAUGCAUCCUGGCCGUCCCGGCCAUCCUGGCCACUUUUCCAGAACUGUUCGUUUAU ...(((((.((.((((((((..(...(((.((((((.....))))))))).)..))))))))))(((((...)))))......))))).......((((.....)))).. ( -46.20) >DroEre_CAF1 85046 92 + 1 GCCGUGGCAAGUGAUGCGUGUGGUAAAUGCUCCAGGACACCCCUGCACAUCCUGCAUGCAUCCU------------------GGCCACUUUUCCAGCACUGUUCGUUUAU ...(((((.((.((((((((..(...(((...((((.....))))..))).)..))))))))))------------------.)))))...................... ( -32.20) >DroYak_CAF1 86147 110 + 1 GCCGUGGCAAGUGAUGCGUGUGGUAAAUGCUGCAGGACACCCCUGCACAUCCUGCAUGCAUCCUGGCCGUCCUGCUCGUCCUGGCCACUUUUCCAGCACUGUUCGUUUAU ((.(((((.((.((((((((..(...(((.((((((.....))))))))).)..)))))))))).)))).)..))..((.((((........)))).))........... ( -40.10) >consensus GCCGUGGCAAGUGAUGCGUGUGGUAAAUGCUGCAGGACACCCCUGCACAUCCUGCAUGCAUCCUGGCCGUC_________CUGGCCACUUUUCCAGCACUGUUCGUUUAU ...(((((.((.((((((((..(...(((.((((((.....))))))))).)..))))))))))...................)))))...................... (-31.70 = -32.20 + 0.50)

| Location | 14,185,650 – 14,185,751 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 88.33 |
| Mean single sequence MFE | -37.83 |
| Consensus MFE | -29.80 |
| Energy contribution | -31.12 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
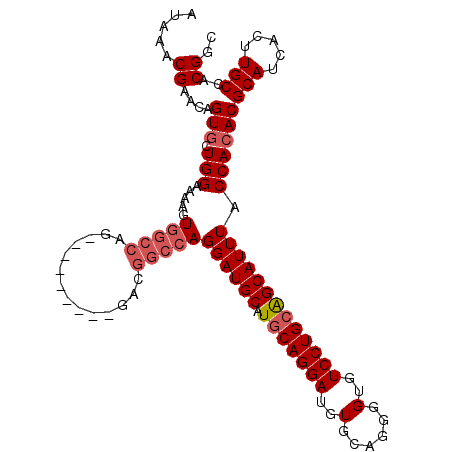
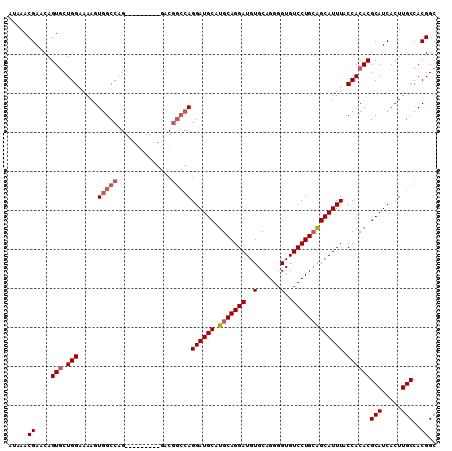
>X_DroMel_CAF1 14185650 101 - 22224390 AUAAACGAACAGUGCUGGAAAAGUGGCCAG---------GACGGCCAGGAUGCAUGCAGGAUGUGCAGGGGUGUCCUGCGGCAUUUACCACACGCAUCACUUGCCACGGC .....((....((.((((........))))---------.))(((.(((((((..((((((..(......)..))))))((......))....))))).)).))).)).. ( -38.10) >DroSec_CAF1 96053 110 - 1 AUAAACGAACAGUUCUGGAAAAGUGGCCAGGAUGGCCGGGACGGCCAGGAUGCAUGCAGGAUGUGCAGGGGUGUCCUGCAGCAUUUACCACACGCAUCACUUGCCACGGC ......................(((((.((((((((((...))))).((((((.(((((((..(......)..))))))))))))).........))).)).)))))... ( -40.80) >DroEre_CAF1 85046 92 - 1 AUAAACGAACAGUGCUGGAAAAGUGGCC------------------AGGAUGCAUGCAGGAUGUGCAGGGGUGUCCUGGAGCAUUUACCACACGCAUCACUUGCCACGGC .....((....(((.(((..(((((.((------------------(((((((.((((.....))))...)))))))))..))))).))))))(((.....)))..)).. ( -32.10) >DroYak_CAF1 86147 110 - 1 AUAAACGAACAGUGCUGGAAAAGUGGCCAGGACGAGCAGGACGGCCAGGAUGCAUGCAGGAUGUGCAGGGGUGUCCUGCAGCAUUUACCACACGCAUCACUUGCCACGGC .....((....(((.(((.....(((((.(..(.....)..))))))((((((.(((((((..(......)..))))))))))))).))))))(((.....)))..)).. ( -40.30) >consensus AUAAACGAACAGUGCUGGAAAAGUGGCCAG_________GACGGCCAGGAUGCAUGCAGGAUGUGCAGGGGUGUCCUGCAGCAUUUACCACACGCAUCACUUGCCACGGC .....((....(((.(((.....(((((..............)))))((((((.(((((((..(......)..))))))))))))).))))))(((.....)))..)).. (-29.80 = -31.12 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:12 2006