
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,184,341 – 14,184,459 |
| Length | 118 |
| Max. P | 0.844871 |

| Location | 14,184,341 – 14,184,459 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 92.26 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -20.70 |
| Energy contribution | -21.20 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
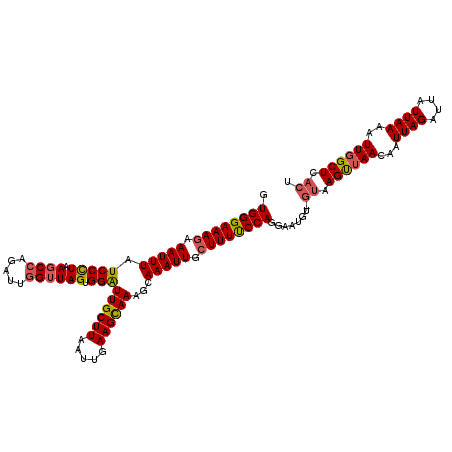
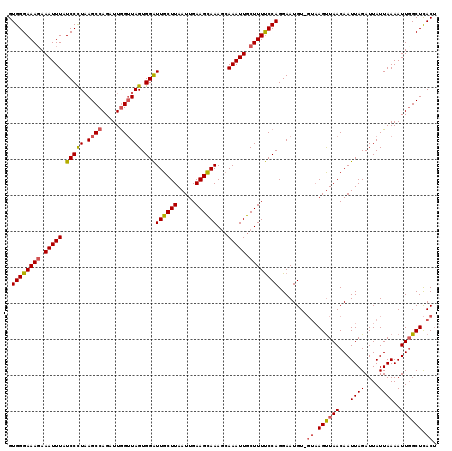
>X_DroMel_CAF1 14184341 118 + 22224390 GUGGGAAAGAAAUUUAUCCCUAAGCCAGAUUGGUUAGUGGAUUGCUUAAUUGAAGUAAAGCAAAUUGCUUUUCCAGGAAUGU-GUAAGCUAACAAUUAGAUUAUUAAAAUUGGCUCACU .((((((((.(((((((((((((.((.....)))))).)))).((((.((....)).))))))))).)))))))).....((-(..((((((...((((....))))..))))))))). ( -27.30) >DroSec_CAF1 94758 117 + 1 GUGGGAAAGAAAUUUAUCCUU-AGCCAGAUUGGUUAGUGGAUUGCUUAAUUGAAGCAAACGAAAUUGCUUUUCCAGGAAUGU-GUAAGUUAACAAUUAGAUUAUUAAAAUUCGCUCACU .((((((((.(((((.(((((-((((.....)))))).)))((((((.....))))))...))))).)))))))).((((..-((((.(((.....))).))))....))))....... ( -27.20) >DroSim_CAF1 72289 118 + 1 GUGGGAAAGAAAUUUAUCCUUAAGCCAGAUUGGUUAGUGGAUUGCUUAAUUGAAGCAAAGCAAAUUGCUUUUCCAGGAAUGU-GUAAGUUAACAAUUAGAUUAUUAAAAUUGGCUCACU .((((((((.(((((.(((...((((.....))))...)))((((((.....))))))...))))).)))))))).....((-(..((((((...((((....))))..))))))))). ( -25.40) >DroYak_CAF1 85018 116 + 1 GUGGGAAACAAAUUUAUCCCUAAGCCAGAUUAGAUAGUGGGUUGCUUAAUUGAAGCAAAGCAAAUUGCUUUCCCAGGAAUGUA---AGUUAACAAUUAGAUUAUUAAAAUUGGCUUACA ..((((..........))))((((((((.((.(((((((((..((((.....))))(((((.....)))))))).....(((.---.....))).....)))))).)).)))))))).. ( -27.90) >consensus GUGGGAAAGAAAUUUAUCCCUAAGCCAGAUUGGUUAGUGGAUUGCUUAAUUGAAGCAAAGCAAAUUGCUUUUCCAGGAAUGU_GUAAGUUAACAAUUAGAUUAUUAAAAUUGGCUCACU .((((((((.(((((.(((((.((((.....)))))).)))((((((.....))))))...))))).))))))))........((.((((((...((((....))))..)))))).)). (-20.70 = -21.20 + 0.50)

| Location | 14,184,341 – 14,184,459 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 92.26 |
| Mean single sequence MFE | -24.05 |
| Consensus MFE | -18.24 |
| Energy contribution | -19.30 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
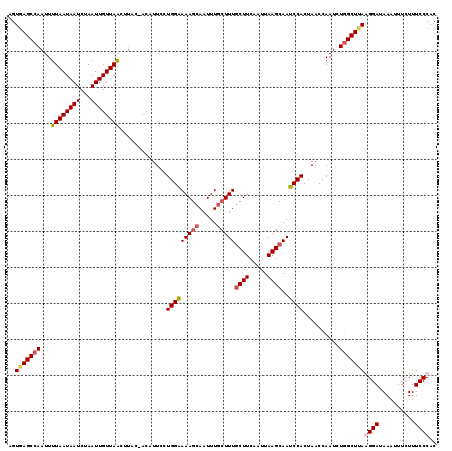
>X_DroMel_CAF1 14184341 118 - 22224390 AGUGAGCCAAUUUUAAUAAUCUAAUUGUUAGCUUAC-ACAUUCCUGGAAAAGCAAUUUGCUUUACUUCAAUUAAGCAAUCCACUAACCAAUCUGGCUUAGGGAUAAAUUUCUUUCCCAC .(((((((((((..........)))))...))))))-.......(((.((((.(((((((((..........)))).((((.((((((.....)).))))))))))))).)))).))). ( -26.20) >DroSec_CAF1 94758 117 - 1 AGUGAGCGAAUUUUAAUAAUCUAAUUGUUAACUUAC-ACAUUCCUGGAAAAGCAAUUUCGUUUGCUUCAAUUAAGCAAUCCACUAACCAAUCUGGCU-AAGGAUAAAUUUCUUUCCCAC .(((((......((((((((...)))))))))))))-.......(((.((((.(((((....(((((.....)))))((((..((.((.....)).)-).))))))))).)))).))). ( -18.50) >DroSim_CAF1 72289 118 - 1 AGUGAGCCAAUUUUAAUAAUCUAAUUGUUAACUUAC-ACAUUCCUGGAAAAGCAAUUUGCUUUGCUUCAAUUAAGCAAUCCACUAACCAAUCUGGCUUAAGGAUAAAUUUCUUUCCCAC ..(((((((...((((((((...)))))))).....-.......(((((((((.....)))))((((.....))))..))))..........))))))).(((..........)))... ( -22.70) >DroYak_CAF1 85018 116 - 1 UGUAAGCCAAUUUUAAUAAUCUAAUUGUUAACU---UACAUUCCUGGGAAAGCAAUUUGCUUUGCUUCAAUUAAGCAACCCACUAUCUAAUCUGGCUUAGGGAUAAAUUUGUUUCCCAC ..(((((((...((((((((...))))))))..---........(((((((((.....)))))((((.....))))..))))..........)))))))((((..........)))).. ( -28.80) >consensus AGUGAGCCAAUUUUAAUAAUCUAAUUGUUAACUUAC_ACAUUCCUGGAAAAGCAAUUUGCUUUGCUUCAAUUAAGCAAUCCACUAACCAAUCUGGCUUAAGGAUAAAUUUCUUUCCCAC ..(((((((...((((((((...)))))))).............(((((((((.....)))))((((.....))))..))))..........)))))))((((..........)))).. (-18.24 = -19.30 + 1.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:10 2006