
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,182,333 – 14,182,471 |
| Length | 138 |
| Max. P | 0.985586 |

| Location | 14,182,333 – 14,182,431 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 91.43 |
| Mean single sequence MFE | -21.29 |
| Consensus MFE | -18.18 |
| Energy contribution | -18.54 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
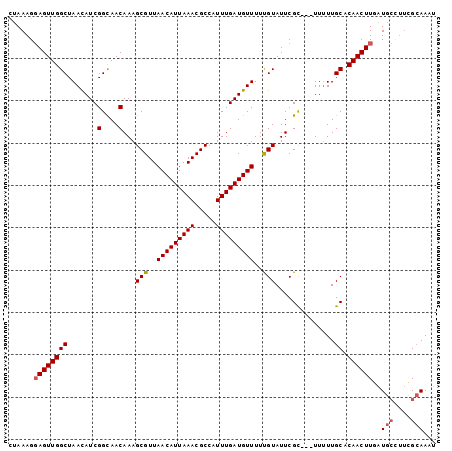
>X_DroMel_CAF1 14182333 98 + 22224390 CUAAAGGAGUUGGCUAACAUCGGCAACAAAGCGUUAACAUUAAACGCCAUUUGAUGUUUUUGUAUUCGU---UUUUUGCACAACUUGAUGCUUUCGCAAAU ......((((((((.(((...(....)...(((..(((((((((.....)))))))))..)))....))---)....)).))))))..(((....)))... ( -21.70) >DroSec_CAF1 92754 98 + 1 CUAAAGGAGUUGGCUAAUAUCGGCAACAAAGCGUUAACAUUAAACGCCAUUUGAUGUUUUUGUAUUCGC---UUUUUGCACAACUUGAUGCCUUCGCAAAU ......((((((((.......(....)((((((..(((((((((.....)))))))))........)))---)))..)).))))))..(((....)))... ( -21.10) >DroSim_CAF1 70270 98 + 1 CUAAAGGAGUUGGCUAACAUCGGCAACAAAGCGUUAACAUUAAACGCCAUUUGAUGUUUUUGUAUUCGU---UUUUUGCACAACUUGAUGCCUUCGCAAAU ......((((((((.(((...(....)...(((..(((((((((.....)))))))))..)))....))---)....)).))))))..(((....)))... ( -21.20) >DroEre_CAF1 82458 92 + 1 UUAAAGCAGUUGGCUAACAUCGGUAACAAAGCGUUAACAUUAAACGCCAUUUGAUGUUUUCGUAUUCGC---UUCUUGCACAACUUGGU------GAAAAU ....(((.....)))..(((((((......(((..(((((((((.....)))))))))..)))....((---.....))...)).))))------)..... ( -18.00) >DroYak_CAF1 83053 101 + 1 UUAAAGGAGUUGGCUAACAUCGGCAACAAAGCGUUAACAUUAAACGCCAUUUGAUGUUUUCGUAUUCGCCUUUUUUUGCACAACUUGAUGCUUUUGCAAAU ......((((((((.......(((...((.(((..(((((((((.....)))))))))..))).)).))).......)).))))))..(((....)))... ( -24.44) >consensus CUAAAGGAGUUGGCUAACAUCGGCAACAAAGCGUUAACAUUAAACGCCAUUUGAUGUUUUUGUAUUCGC___UUUUUGCACAACUUGAUGCCUUCGCAAAU ......((((((((.......(....)...(((..(((((((((.....)))))))))..)))..............)).))))))..(((....)))... (-18.18 = -18.54 + 0.36)


| Location | 14,182,357 – 14,182,471 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 88.66 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -20.80 |
| Energy contribution | -22.08 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.904149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
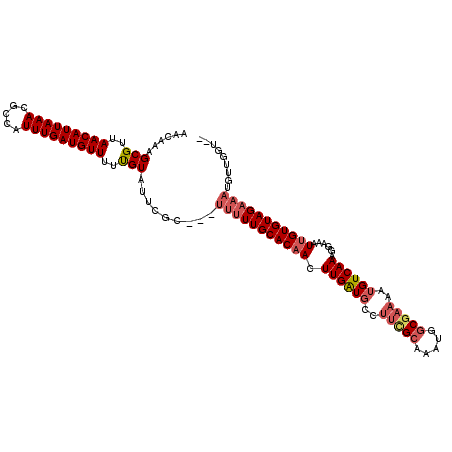
>X_DroMel_CAF1 14182357 114 + 22224390 AACAAAGCGUUAACAUUAAACGCCAUUUGAUGUUUUUGUAUUCGU---UUUUUGCACAACUUGAUGCUUUCGCAAAUGGCGAAAAUGUCAAAGGAAAUUGUGUAGAAAUGUUGGUGA ((((..(((..(((((((((.....)))))))))..)))......---(((((((((((((((((..((((((.....))))))..))).)))....)))))))))))))))..... ( -30.40) >DroSec_CAF1 92778 112 + 1 AACAAAGCGUUAACAUUAAACGCCAUUUGAUGUUUUUGUAUUCGC---UUUUUGCACAACUUGAUGCCUUCGCAAAUGGCGAAAAUGUCAAAGGAAAUAGUGUAGAAAUUUUGGU-- ..(((((..((.((((((....((.((((((((((((((....((---.....)).........(((....)))....))))))))))))))))...)))))).))..)))))..-- ( -27.40) >DroSim_CAF1 70294 112 + 1 AACAAAGCGUUAACAUUAAACGCCAUUUGAUGUUUUUGUAUUCGU---UUUUUGCACAACUUGAUGCCUUCGCAAAUGGCGAAAAUGUCAAAGGAAAUUGUGUAGAAAUUUUGGU-- ..(((((((..(((((((((.....)))))))))..)))......---((((((((((((((((((..(((((.....)))))..)))).)))....))))))))))).))))..-- ( -28.10) >DroEre_CAF1 82482 97 + 1 AACAAAGCGUUAACAUUAAACGCCAUUUGAUGUUUUCGUAUUCGC---UUCUUGCACAACUUGGU------GAAAAUGGCGAAAAUGUCAAACGAAAUUGUGUAGA----------- .((((.(((((.......)))))..((((((((((((((.(((((---(.............)))------)))....)))))))))))))).....)))).....----------- ( -25.02) >DroYak_CAF1 83077 117 + 1 AACAAAGCGUUAACAUUAAACGCCAUUUGAUGUUUUCGUAUUCGCCUUUUUUUGCACAACUUGAUGCUUUUGCAAAUGGCGAAAAUGUCAAAGGAAAUUGUGUAGAAAUGUUGAUGA .......((((((((((..((((..(((((((((((((....)(((....((((((.(((.....).)).)))))).))))))))))))))).......))))...)))))))))). ( -30.20) >consensus AACAAAGCGUUAACAUUAAACGCCAUUUGAUGUUUUUGUAUUCGC___UUUUUGCACAACUUGAUGCCUUCGCAAAUGGCGAAAAUGUCAAAGGAAAUUGUGUAGAAAUGUUGGU__ ......(((..(((((((((.....)))))))))..))).........(((((((((((.((((((..(((((.....)))))..))))))......)))))))))))......... (-20.80 = -22.08 + 1.28)


| Location | 14,182,357 – 14,182,471 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 88.66 |
| Mean single sequence MFE | -23.82 |
| Consensus MFE | -19.22 |
| Energy contribution | -20.02 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14182357 114 - 22224390 UCACCAACAUUUCUACACAAUUUCCUUUGACAUUUUCGCCAUUUGCGAAAGCAUCAAGUUGUGCAAAAA---ACGAAUACAAAAACAUCAAAUGGCGUUUAAUGUUAACGCUUUGUU ................((((....(.((((((((..((((((((((....))......((((.......---)))).............))))))))...)))))))).)..)))). ( -24.50) >DroSec_CAF1 92778 112 - 1 --ACCAAAAUUUCUACACUAUUUCCUUUGACAUUUUCGCCAUUUGCGAAGGCAUCAAGUUGUGCAAAAA---GCGAAUACAAAAACAUCAAAUGGCGUUUAAUGUUAACGCUUUGUU --........................((((((((..((((((((((....)).....((..(((.....---)))...)).........))))))))...))))))))......... ( -24.60) >DroSim_CAF1 70294 112 - 1 --ACCAAAAUUUCUACACAAUUUCCUUUGACAUUUUCGCCAUUUGCGAAGGCAUCAAGUUGUGCAAAAA---ACGAAUACAAAAACAUCAAAUGGCGUUUAAUGUUAACGCUUUGUU --..............((((....(.((((((((..((((((((((....))......((((.......---)))).............))))))))...)))))))).)..)))). ( -23.40) >DroEre_CAF1 82482 97 - 1 -----------UCUACACAAUUUCGUUUGACAUUUUCGCCAUUUUC------ACCAAGUUGUGCAAGAA---GCGAAUACGAAAACAUCAAAUGGCGUUUAAUGUUAACGCUUUGUU -----------.....((((...((((.((((((..((((((((..------.....((..(((.....---)))...)).........))))))))...))))))))))..)))). ( -22.99) >DroYak_CAF1 83077 117 - 1 UCAUCAACAUUUCUACACAAUUUCCUUUGACAUUUUCGCCAUUUGCAAAAGCAUCAAGUUGUGCAAAAAAAGGCGAAUACGAAAACAUCAAAUGGCGUUUAAUGUUAACGCUUUGUU ................((((....(.((((((((..(((((((((.....((((......)))).........((....)).......)))))))))...)))))))).)..)))). ( -23.60) >consensus __ACCAAAAUUUCUACACAAUUUCCUUUGACAUUUUCGCCAUUUGCGAAAGCAUCAAGUUGUGCAAAAA___GCGAAUACAAAAACAUCAAAUGGCGUUUAAUGUUAACGCUUUGUU ..........................((((((((..((((((((((....)).....((..(((........)))...)).........))))))))...))))))))......... (-19.22 = -20.02 + 0.80)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:06 2006