
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,179,341 – 14,179,436 |
| Length | 95 |
| Max. P | 0.954017 |

| Location | 14,179,341 – 14,179,436 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 78.28 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -15.23 |
| Energy contribution | -15.03 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
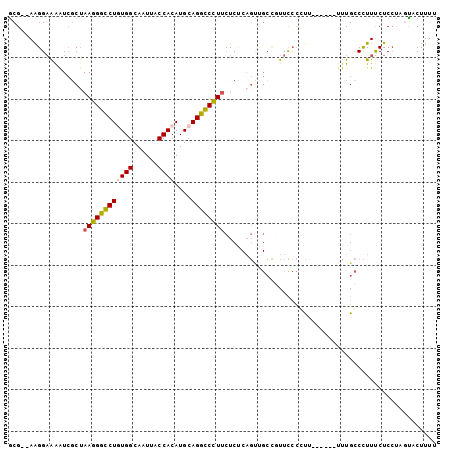
>X_DroMel_CAF1 14179341 95 + 22224390 ACG--AAGGAAAAUCGCUAAGGGCCUAUGGCAAUUACCACAUGCAGGCCCUUCUCUCAGUUGCCGUUUCCGUU------UUUGCCCUUUCUCCUAGUACUUUU ...--..(((((...((.((((((((.(((......))).....)))))))).........))..)))))...------........................ ( -19.50) >DroSec_CAF1 89765 95 + 1 GCG--AAGGAAAAUCGCGAAGGGCCUGUGGCAAUUACCACAUGCAGGUCCUUCUCCCAGUUGCCGUUCCCGUU------AUUGCCCUUUCUCCUAGUACUUUU ((.--.((((.......(((((((((((((......))....)))))))))))...((((.((.......)).------)))).......)))).))...... ( -24.20) >DroSim_CAF1 67288 95 + 1 GCG--AAGGAAAAUCGCUAAGGGCCUGUGGCAAUUACCACAUGCAGGCCCUUCUCUCAGUUGCCGUUCCCGUU------UUUGCCCUUUCUCCUAGUACUUUU (((--((((((....(((((((((((((((......))....)))))))))).....))).....))))....------)))))................... ( -24.60) >DroEre_CAF1 79574 94 + 1 GCG--AAGGAAAAUCGCUAAGGGCCUGUGGCAAUUACCACAUGCAGGCCCUUCUCUCAGCCGCU-UUCCCCUC------UUUGCCCUUCCUCCCAUUACUUUU (((--(((((((...(((((((((((((((......))....)))))))))).....)))...)-))))....------)))))................... ( -26.90) >DroYak_CAF1 80205 100 + 1 GCG--AAGGAAAAUCACUAAGGGCCUGUGGCAAUGACCACAUGCAGGCCCUUCUCUCAGCCGCU-UUCCCCUUUUUUUUUUUGCCCUUUCUCCCAUUACUUUU (((--((((((((.....((((((.(((((......))))).)).(((..........)))...-....)))))))))))))))................... ( -23.70) >DroAna_CAF1 85380 85 + 1 GCGAUAGGGCGGGUAGCUAAAGGGCUCUGGCAAUUACCACAUGAAGUCCUUCGUC---------CUUCAUCCU------UCUGGCUUUCCUC---GCACUUUA .....((((((((.(((((((((((....)).........((((((..(...)..---------)))))))))------).)))))...)))---)).))).. ( -22.60) >consensus GCG__AAGGAAAAUCGCUAAGGGCCUGUGGCAAUUACCACAUGCAGGCCCUUCUCUCAGUUGCCGUUCCCCUU______UUUGCCCUUUCUCCUAGUACUUUU ..................((((((((((((......))))....))))))))................................................... (-15.23 = -15.03 + -0.19)


| Location | 14,179,341 – 14,179,436 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 78.28 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -20.53 |
| Energy contribution | -20.26 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
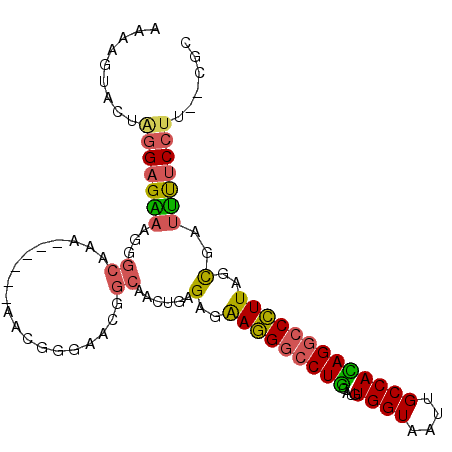
>X_DroMel_CAF1 14179341 95 - 22224390 AAAAGUACUAGGAGAAAGGGCAAA------AACGGAAACGGCAACUGAGAGAAGGGCCUGCAUGUGGUAAUUGCCAUAGGCCCUUAGCGAUUUUCCUU--CGU .........(((((((...((...------..((....))...........((((((((....(((((....))))))))))))).))..))))))).--... ( -30.70) >DroSec_CAF1 89765 95 - 1 AAAAGUACUAGGAGAAAGGGCAAU------AACGGGAACGGCAACUGGGAGAAGGACCUGCAUGUGGUAAUUGCCACAGGCCCUUCGCGAUUUUCCUU--CGC .............(((.((((...------..((....))(((...((........))))).((((((....)))))).)))))))((((.......)--))) ( -27.70) >DroSim_CAF1 67288 95 - 1 AAAAGUACUAGGAGAAAGGGCAAA------AACGGGAACGGCAACUGAGAGAAGGGCCUGCAUGUGGUAAUUGCCACAGGCCCUUAGCGAUUUUCCUU--CGC .........(((((((...((...------..((....))...........((((((((....(((((....))))))))))))).))..))))))).--... ( -30.50) >DroEre_CAF1 79574 94 - 1 AAAAGUAAUGGGAGGAAGGGCAAA------GAGGGGAA-AGCGGCUGAGAGAAGGGCCUGCAUGUGGUAAUUGCCACAGGCCCUUAGCGAUUUUCCUU--CGC ..........(((((((((.....------(((((...-.((((((........)))).)).((((((....))))))..))))).....))))))))--).. ( -33.50) >DroYak_CAF1 80205 100 - 1 AAAAGUAAUGGGAGAAAGGGCAAAAAAAAAAAGGGGAA-AGCGGCUGAGAGAAGGGCCUGCAUGUGGUCAUUGCCACAGGCCCUUAGUGAUUUUCCUU--CGC .........((((((((((((.................-.((((((........)))).)).((((((....)))))).)))))).....))))))..--... ( -28.90) >DroAna_CAF1 85380 85 - 1 UAAAGUGC---GAGGAAAGCCAGA------AGGAUGAAG---------GACGAAGGACUUCAUGUGGUAAUUGCCAGAGCCCUUUAGCUACCCGCCCUAUCGC ...((.((---(.((..(((..((------(((((((((---------..(....).)))))).((((....))))....))))).))).))))).))..... ( -24.50) >consensus AAAAGUACUAGGAGAAAGGGCAAA______AACGGGAACGGCAACUGAGAGAAGGGCCUGCAUGUGGUAAUUGCCACAGGCCCUUAGCGAUUUUCCUU__CGC .........(((((((...((...................))......(..(((((((((....((((....)))))))))))))..)..)))))))...... (-20.53 = -20.26 + -0.27)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:00 2006