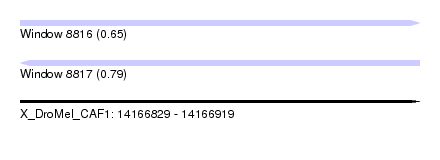
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,166,829 – 14,166,919 |
| Length | 90 |
| Max. P | 0.785925 |

| Location | 14,166,829 – 14,166,919 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 87.43 |
| Mean single sequence MFE | -34.85 |
| Consensus MFE | -29.00 |
| Energy contribution | -30.62 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
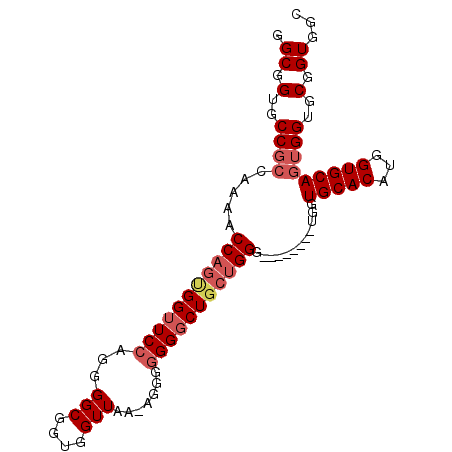
>X_DroMel_CAF1 14166829 90 + 22224390 GGCGGUGCC------ACCAGUGGUUCCAAGGGCGGUGGUUAACAGGGGGGGCUGCUGGGUGGUUGGGUGGUGCACAUGGUGCAGUGGUGCGGUGGC .((.(..((------(((((..(((((.......((.....))....)))))..))))............(((((...))))).)))..).))... ( -32.60) >DroSec_CAF1 77634 87 + 1 GGCGGUGCCGCCAAAACCAGCGGUUCCAGGGGCGGUGGUUAA-AGGGGGGGCUGCUGGG--------UGGUGCACAUGGUGCAGUGGUGCGGUGGC .((.(..((((((...(((((((((((...(((....)))..-....))))))))))).--------))))((((...))))...))..).))... ( -39.50) >DroSim_CAF1 55220 87 + 1 GGCGGUGCCGCCAAAACCAGUGGUUCCAGGGGCGGUGGUUAA-AGGGGGGGCUGCUGGG--------UGGUGCACAUGGUGCAGUGGUGCGGUGGC .((.(..((((((...((((..(((((...(((....)))..-....)))))..)))).--------))))((((...))))...))..).))... ( -37.00) >DroYak_CAF1 67833 83 + 1 GGCGGUGCCGCCAAAACCAGUGGUUCCAGGGGCGGUGGUUAA-AGG--GGUCUU--GGG--------UGGUGCACAUGGUGCAGUGGUGCGGUGGC .((.(..((((((...((((.((..((...(((....)))..-.))--..))))--)).--------))))((((...))))...))..).))... ( -30.30) >consensus GGCGGUGCCGCCAAAACCAGUGGUUCCAGGGGCGGUGGUUAA_AGGGGGGGCUGCUGGG________UGGUGCACAUGGUGCAGUGGUGCGGUGGC .((.(..((((.....(((((((((((...(((....))).......)))))))))))............(((((...)))))))))..).))... (-29.00 = -30.62 + 1.62)

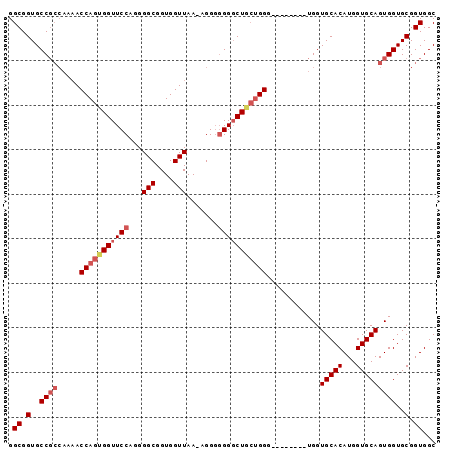
| Location | 14,166,829 – 14,166,919 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 87.43 |
| Mean single sequence MFE | -21.93 |
| Consensus MFE | -16.62 |
| Energy contribution | -17.38 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
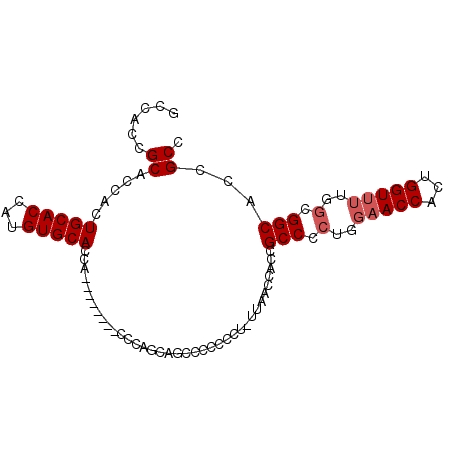
>X_DroMel_CAF1 14166829 90 - 22224390 GCCACCGCACCACUGCACCAUGUGCACCACCCAACCACCCAGCAGCCCCCCCUGUUAACCACCGCCCUUGGAACCACUGGU------GGCACCGCC ((((((((.....(((((...)))))..............(((((......))))).......))...((....))..)))------)))...... ( -21.90) >DroSec_CAF1 77634 87 - 1 GCCACCGCACCACUGCACCAUGUGCACCA--------CCCAGCAGCCCCCCCU-UUAACCACCGCCCCUGGAACCGCUGGUUUUGGCGGCACCGCC (((.(((.((((.(((((...)))))...--------.((((..((.......-.........))..))))......))))..))).)))...... ( -22.29) >DroSim_CAF1 55220 87 - 1 GCCACCGCACCACUGCACCAUGUGCACCA--------CCCAGCAGCCCCCCCU-UUAACCACCGCCCCUGGAACCACUGGUUUUGGCGGCACCGCC (((.(((.((((.(((((...)))))...--------.((((..((.......-.........))..))))......))))..))).)))...... ( -22.29) >DroYak_CAF1 67833 83 - 1 GCCACCGCACCACUGCACCAUGUGCACCA--------CCC--AAGACC--CCU-UUAACCACCGCCCCUGGAACCACUGGUUUUGGCGGCACCGCC (((..........(((((...)))))...--------.((--((((((--...-....(((.......))).......)))))))).)))...... ( -21.24) >consensus GCCACCGCACCACUGCACCAUGUGCACCA________CCCAGCAGCCCCCCCU_UUAACCACCGCCCCUGGAACCACUGGUUUUGGCGGCACCGCC ......((.....(((((...))))).....................................(((.(..(((((...)))))..).)))...)). (-16.62 = -17.38 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:52 2006