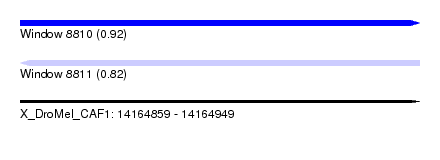
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,164,859 – 14,164,949 |
| Length | 90 |
| Max. P | 0.921933 |

| Location | 14,164,859 – 14,164,949 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 70.48 |
| Mean single sequence MFE | -22.44 |
| Consensus MFE | -3.56 |
| Energy contribution | -3.84 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.16 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14164859 90 + 22224390 UAAUA------UUUAUUUUAUGUAUCUCAGGUUGGCCUUGAAAGUAAGGUGCUAAUGAAAAUUGGAGCUGAUAAGUUUGACAACCCCAAUGCA--CCC .....------.........(((((....(((((((((((....)))))).(((((....)))))((((....))))...)))))...)))))--... ( -18.70) >DroSec_CAF1 75439 96 + 1 UAACAUUGGGAUUUAUUUAAUGUAUCUCCAAAUGGCCUUGAAAAUAAGGUGCCAAUGAAAAUUGGAGAUGAUAAGUUUGACAAUCCCAAUGCA--CCC ...((((((((((.(((((...(((((((((.(((((((.......))).)))).(....)))))))))).))))).....))))))))))..--... ( -30.70) >DroSim_CAF1 53292 96 + 1 UAAUAUUGGGAUUUAUUUAAUGUAUCUCAAAAUGGCCUUGAAAAUAAGGUGCCAAUGAAAAUUGGAGAUGAUAAGUUGACAAAUCCCAAUGCA--CCC ...(((((((((((..(((((.((((((......((((((....)))))).(((((....))))).)).)))).))))).)))))))))))..--... ( -29.90) >DroEre_CAF1 65143 85 + 1 -----------UCUGUGGAAGAUAUUGUCAAUUGGCCCUGAAAAUAAGGUGCUAAUGAAAACGGGUUAAGUUAAGCUAGAUAGCCCCAAUGCA--CCC -----------.....((..(.((((((((.((((((((.......))).))))))))....((((((((.....))...)))))))))))).--.)) ( -19.00) >DroYak_CAF1 65923 87 + 1 -----------UCUGUGGGAAAAGUGGUCGAAUGGCCUUGAAAUUAAGAUGGAAAUGAAAACAGGAGCCGAUAGGUUAGACAACCCAAAUGCAACCCC -----------....((((......((((....))))..........................(.((((....))))...)..))))........... ( -13.90) >consensus UAA_A______UUUAUUUAAUGUAUCUCCAAAUGGCCUUGAAAAUAAGGUGCCAAUGAAAAUUGGAGAUGAUAAGUUAGACAACCCCAAUGCA__CCC ................................(((((((.......))).))))............................................ ( -3.56 = -3.84 + 0.28)

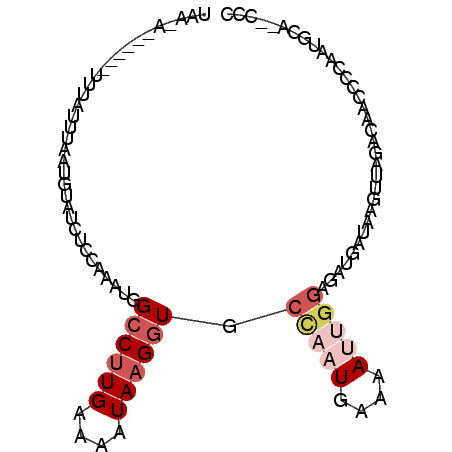
| Location | 14,164,859 – 14,164,949 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 70.48 |
| Mean single sequence MFE | -20.47 |
| Consensus MFE | -4.58 |
| Energy contribution | -5.02 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.22 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14164859 90 - 22224390 GGG--UGCAUUGGGGUUGUCAAACUUAUCAGCUCCAAUUUUCAUUAGCACCUUACUUUCAAGGCCAACCUGAGAUACAUAAAAUAAA------UAUUA (((--(((((((((((((..........))))))))))........))))))...(((((.((....))))))).............------..... ( -22.40) >DroSec_CAF1 75439 96 - 1 GGG--UGCAUUGGGAUUGUCAAACUUAUCAUCUCCAAUUUUCAUUGGCACCUUAUUUUCAAGGCCAUUUGGAGAUACAUUAAAUAAAUCCCAAUGUUA ...--.(((((((((((............((((((((.......((((..(((......))))))).))))))))..........))))))))))).. ( -27.75) >DroSim_CAF1 53292 96 - 1 GGG--UGCAUUGGGAUUUGUCAACUUAUCAUCUCCAAUUUUCAUUGGCACCUUAUUUUCAAGGCCAUUUUGAGAUACAUUAAAUAAAUCCCAAUAUUA ...--...((((((((((((.....((((.((.(((((....)))))..((((......)))).......))))))......)))))))))))).... ( -21.00) >DroEre_CAF1 65143 85 - 1 GGG--UGCAUUGGGGCUAUCUAGCUUAACUUAACCCGUUUUCAUUAGCACCUUAUUUUCAGGGCCAAUUGACAAUAUCUUCCACAGA----------- (((--(......(((((....)))))......))))(((...(((.((.(((.......))))).))).)))...............----------- ( -17.10) >DroYak_CAF1 65923 87 - 1 GGGGUUGCAUUUGGGUUGUCUAACCUAUCGGCUCCUGUUUUCAUUUCCAUCUUAAUUUCAAGGCCAUUCGACCACUUUUCCCACAGA----------- ..(((((....((((((....))))))..((((..((.....................)).))))...)))))..............----------- ( -14.10) >consensus GGG__UGCAUUGGGGUUGUCAAACUUAUCAUCUCCAAUUUUCAUUAGCACCUUAUUUUCAAGGCCAUUUGGAGAUACAUUAAAUAAA______U_UUA (((.........(((((....)))))......))).(((((((......((((......)))).....)))))))....................... ( -4.58 = -5.02 + 0.44)

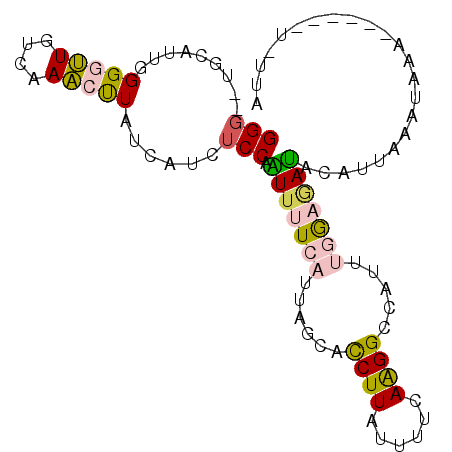
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:46 2006