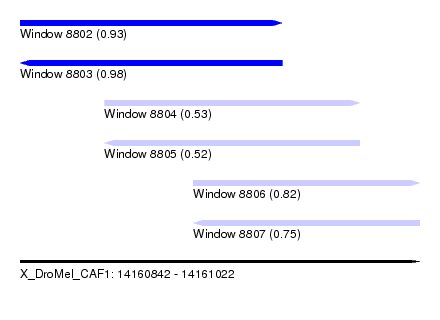
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,160,842 – 14,161,022 |
| Length | 180 |
| Max. P | 0.976967 |

| Location | 14,160,842 – 14,160,960 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.07 |
| Mean single sequence MFE | -33.25 |
| Consensus MFE | -30.85 |
| Energy contribution | -31.52 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14160842 118 + 22224390 -CCCGCAUCCAUUUAGAAGGCAUCCUGACCGGAA-AGACCGCGAGGACAACGGGAAAAGCAAUAAACUUGUUUGUAUUUGUAUUGCACUUAUGCGCUGAUUGCUUUUCAGCGCAAUUUGC -((((..(((..((((........))))..))).-...((....))....))))....((((((.((......)).....)))))).....((((((((.......))))))))...... ( -33.80) >DroSec_CAF1 68937 117 + 1 --CCGCAUCCAUUUAGAAGGCAUCCUGACCGGAA-AGACCGCGAGGACAACGGGAAAGGCAAUAAACUUGUUUGUAUUUGUAUUGCACUUAUGCGCUGAUUGCUUUUCAGCGCAAUUUGC --(((..(((..((((........))))..))).-...((....))....)))..(((((((((.((......)).....)))))).))).((((((((.......))))))))...... ( -31.80) >DroSim_CAF1 48042 118 + 1 -CCCGCAUCCAUUUAGAAGGCAUCCUGACCGGAA-AGACCGCGAGGACAACGGGAAAAGCAAUAAACUUGUUUGUAUUUGUAUUGCACUUAUGCGCUGAUUGCUUUUCAGCGCAAUUUGC -((((..(((..((((........))))..))).-...((....))....))))....((((((.((......)).....)))))).....((((((((.......))))))))...... ( -33.80) >DroEre_CAF1 61148 118 + 1 -CCCGCAUCCAUUUAGAAGGCUUCCUGACCGGAA-AGACCGCGAGGACAACGGGAAAAGCAAUAAACUUGUUUGUAUUUGUAUUGCACUUAUGCGCUGAUUGCUUUUCAGCGCAAUUUGC -((((..(((..((((........))))..))).-...((....))....))))....((((((.((......)).....)))))).....((((((((.......))))))))...... ( -33.80) >DroYak_CAF1 61953 119 + 1 CCCCGCAUCCAUUUAGAAGGCAUCCUGACCGGAA-AGACCGCGAGGACAACGGGAAAAGCAAUAAACUUGUUUGUAUUUGUAUUGCACUUAUGCGCUGAUUGCUUUUCAGCGCAAUUUGC .((((..(((..((((........))))..))).-...((....))....))))....((((((.((......)).....)))))).....((((((((.......))))))))...... ( -34.10) >DroAna_CAF1 64957 117 + 1 -CCCGCG--AAUUUAGCAUUUAUCCUGACCGGAAAAAACCGCGAGGACAACGGGAAAAGCAAUAAACUUGUUUGUAUUUGUAUUGCACUUAUGCGCUGAUUGCUUUUCAGCGCAAUUUGC -(((((.--......)).....((((...(((......)))..))))....)))....((((((.((......)).....)))))).....((((((((.......))))))))...... ( -32.20) >consensus _CCCGCAUCCAUUUAGAAGGCAUCCUGACCGGAA_AGACCGCGAGGACAACGGGAAAAGCAAUAAACUUGUUUGUAUUUGUAUUGCACUUAUGCGCUGAUUGCUUUUCAGCGCAAUUUGC .((((..(((..((((........))))..))).....((....))....))))....((((((.((......)).....)))))).....((((((((.......))))))))...... (-30.85 = -31.52 + 0.67)



| Location | 14,160,842 – 14,160,960 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.07 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -31.48 |
| Energy contribution | -31.57 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14160842 118 - 22224390 GCAAAUUGCGCUGAAAAGCAAUCAGCGCAUAAGUGCAAUACAAAUACAAACAAGUUUAUUGCUUUUCCCGUUGUCCUCGCGGUCU-UUCCGGUCAGGAUGCCUUCUAAAUGGAUGCGGG- (((...((((((((.......))))))))....)))...............((((.....))))..(((((.(((((.((((...-..)).)).))))).((........))..)))))- ( -33.80) >DroSec_CAF1 68937 117 - 1 GCAAAUUGCGCUGAAAAGCAAUCAGCGCAUAAGUGCAAUACAAAUACAAACAAGUUUAUUGCCUUUCCCGUUGUCCUCGCGGUCU-UUCCGGUCAGGAUGCCUUCUAAAUGGAUGCGG-- (((...((((((((.......)))))))).(((.((((((.....((......)).)))))))))..((((((((((.((((...-..)).)).)))))........))))).)))..-- ( -30.60) >DroSim_CAF1 48042 118 - 1 GCAAAUUGCGCUGAAAAGCAAUCAGCGCAUAAGUGCAAUACAAAUACAAACAAGUUUAUUGCUUUUCCCGUUGUCCUCGCGGUCU-UUCCGGUCAGGAUGCCUUCUAAAUGGAUGCGGG- (((...((((((((.......))))))))....)))...............((((.....))))..(((((.(((((.((((...-..)).)).))))).((........))..)))))- ( -33.80) >DroEre_CAF1 61148 118 - 1 GCAAAUUGCGCUGAAAAGCAAUCAGCGCAUAAGUGCAAUACAAAUACAAACAAGUUUAUUGCUUUUCCCGUUGUCCUCGCGGUCU-UUCCGGUCAGGAAGCCUUCUAAAUGGAUGCGGG- (((...((((((((.......))))))))....)))...............((((.....))))..(((((.((((....(((..-((((.....)))))))........)))))))))- ( -33.60) >DroYak_CAF1 61953 119 - 1 GCAAAUUGCGCUGAAAAGCAAUCAGCGCAUAAGUGCAAUACAAAUACAAACAAGUUUAUUGCUUUUCCCGUUGUCCUCGCGGUCU-UUCCGGUCAGGAUGCCUUCUAAAUGGAUGCGGGG (((...((((((((.......))))))))....)))...............((((.....))))..(((((.(((((.((((...-..)).)).))))).((........))..))))). ( -35.10) >DroAna_CAF1 64957 117 - 1 GCAAAUUGCGCUGAAAAGCAAUCAGCGCAUAAGUGCAAUACAAAUACAAACAAGUUUAUUGCUUUUCCCGUUGUCCUCGCGGUUUUUUCCGGUCAGGAUAAAUGCUAAAUU--CGCGGG- (((...((((((((.......))))))))....)))...............((((.....))))..(((.(((((((.((((......)).)).)))))))..((......--.)))))- ( -32.90) >consensus GCAAAUUGCGCUGAAAAGCAAUCAGCGCAUAAGUGCAAUACAAAUACAAACAAGUUUAUUGCUUUUCCCGUUGUCCUCGCGGUCU_UUCCGGUCAGGAUGCCUUCUAAAUGGAUGCGGG_ (((...((((((((.......))))))))....)))..............................(((((((((((.((((......)).)).))))))((........))..))))). (-31.48 = -31.57 + 0.09)



| Location | 14,160,880 – 14,160,995 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.17 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -24.74 |
| Energy contribution | -24.86 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14160880 115 + 22224390 GCGAGGACAACGGGAAAAGCAAUAAACUUGUUUGUAUUUGUAUUGCACUUAUGCGCUGAUUGCUUUUCAGCGCAAUUUGCUUUUAAGCGAUUCCUAGUGAGGACUGUCGGUUUUU----- ..((((((......((((((((..........((((.......))))....((((((((.......))))))))..))))))))...((((((((....))))..))))))))))----- ( -31.70) >DroSec_CAF1 68974 115 + 1 GCGAGGACAACGGGAAAGGCAAUAAACUUGUUUGUAUUUGUAUUGCACUUAUGCGCUGAUUGCUUUUCAGCGCAAUUUGCUUUUAAGCGAUUCCUAGUGAGGACUGUCGGUUUUU----- ..((((((.((((..(((((((((.((......)).....)))))).))).((((((((.......))))))))..(((((....))))).((((....))))))))..))))))----- ( -31.60) >DroEre_CAF1 61186 115 + 1 GCGAGGACAACGGGAAAAGCAAUAAACUUGUUUGUAUUUGUAUUGCACUUAUGCGCUGAUUGCUUUUCAGCGCAAUUUGCUUUUAAGCGAUUCCUAGUGAGGACUGUCGGUUUUC----- ..((((((......((((((((..........((((.......))))....((((((((.......))))))))..))))))))...((((((((....))))..))))))))))----- ( -33.30) >DroWil_CAF1 50320 92 + 1 GAGAGAAA-----AAAAACCAAUAAACUUGUUUGUAUUUGUAUUGCACUUAUGCGCUGAUUGCUUUUCAGCGCAAUUUGCUUUUAAGCGAUUUCAGC----------------------- ..((((..-----...................((((.......))))....((((((((.......))))))))..(((((....)))))))))...----------------------- ( -19.70) >DroYak_CAF1 61992 115 + 1 GCGAGGACAACGGGAAAAGCAAUAAACUUGUUUGUAUUUGUAUUGCACUUAUGCGCUGAUUGCUUUUCAGCGCAAUUUGCUUUUAAGCGAUUCCUAGUGAGGACUGUCGGUUUUC----- ..((((((......((((((((..........((((.......))))....((((((((.......))))))))..))))))))...((((((((....))))..))))))))))----- ( -33.30) >DroAna_CAF1 64994 120 + 1 GCGAGGACAACGGGAAAAGCAAUAAACUUGUUUGUAUUUGUAUUGCACUUAUGCGCUGAUUGCUUUUCAGCGCAAUUUGCUUUUAAGUGAUUCUUAGUAGAGACCAUCCCUCUACUACCA ..(((((..((...((((((((..........((((.......))))....((((((((.......))))))))..))))))))..))..)))))(((((((.......))))))).... ( -36.40) >consensus GCGAGGACAACGGGAAAAGCAAUAAACUUGUUUGUAUUUGUAUUGCACUUAUGCGCUGAUUGCUUUUCAGCGCAAUUUGCUUUUAAGCGAUUCCUAGUGAGGACUGUCGGUUUUC_____ ((.((((.......((((((((..........((((.......))))....((((((((.......))))))))..)))))))).......)))).))...................... (-24.74 = -24.86 + 0.11)

| Location | 14,160,880 – 14,160,995 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.17 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -19.63 |
| Energy contribution | -19.86 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14160880 115 - 22224390 -----AAAAACCGACAGUCCUCACUAGGAAUCGCUUAAAAGCAAAUUGCGCUGAAAAGCAAUCAGCGCAUAAGUGCAAUACAAAUACAAACAAGUUUAUUGCUUUUCCCGUUGUCCUCGC -----.......(((((((((....))))..((...((((((((..((((((((.......))))))))...(((...))).................))))))))..)))))))..... ( -25.60) >DroSec_CAF1 68974 115 - 1 -----AAAAACCGACAGUCCUCACUAGGAAUCGCUUAAAAGCAAAUUGCGCUGAAAAGCAAUCAGCGCAUAAGUGCAAUACAAAUACAAACAAGUUUAUUGCCUUUCCCGUUGUCCUCGC -----.......(((((((((....))))...(((....)))....((((((((.......)))))))).(((.((((((.....((......)).))))))))).....)))))..... ( -25.50) >DroEre_CAF1 61186 115 - 1 -----GAAAACCGACAGUCCUCACUAGGAAUCGCUUAAAAGCAAAUUGCGCUGAAAAGCAAUCAGCGCAUAAGUGCAAUACAAAUACAAACAAGUUUAUUGCUUUUCCCGUUGUCCUCGC -----.......(((((((((....))))..((...((((((((..((((((((.......))))))))...(((...))).................))))))))..)))))))..... ( -25.60) >DroWil_CAF1 50320 92 - 1 -----------------------GCUGAAAUCGCUUAAAAGCAAAUUGCGCUGAAAAGCAAUCAGCGCAUAAGUGCAAUACAAAUACAAACAAGUUUAUUGGUUUUU-----UUUCUCUC -----------------------...((((((........(((...((((((((.......))))))))....))).....((((........))))...)))))).-----........ ( -18.90) >DroYak_CAF1 61992 115 - 1 -----GAAAACCGACAGUCCUCACUAGGAAUCGCUUAAAAGCAAAUUGCGCUGAAAAGCAAUCAGCGCAUAAGUGCAAUACAAAUACAAACAAGUUUAUUGCUUUUCCCGUUGUCCUCGC -----.......(((((((((....))))..((...((((((((..((((((((.......))))))))...(((...))).................))))))))..)))))))..... ( -25.60) >DroAna_CAF1 64994 120 - 1 UGGUAGUAGAGGGAUGGUCUCUACUAAGAAUCACUUAAAAGCAAAUUGCGCUGAAAAGCAAUCAGCGCAUAAGUGCAAUACAAAUACAAACAAGUUUAUUGCUUUUCCCGUUGUCCUCGC .(((((((((((.....)))))))))..........((((((((..((((((((.......))))))))...(((...))).................)))))))).))........... ( -30.00) >consensus _____GAAAACCGACAGUCCUCACUAGGAAUCGCUUAAAAGCAAAUUGCGCUGAAAAGCAAUCAGCGCAUAAGUGCAAUACAAAUACAAACAAGUUUAUUGCUUUUCCCGUUGUCCUCGC ..........................(((.......((((((((..((((((((.......))))))))...(((...))).................)))))))).......))).... (-19.63 = -19.86 + 0.22)



| Location | 14,160,920 – 14,161,022 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.19 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -21.65 |
| Energy contribution | -21.27 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14160920 102 + 22224390 UAUUGCACUUAUGCGCUGAUUGCUUUUCAGCGCAAUUUGCUUUUAAGCGAUUCCUAGUGAGGACUGUCGGUUUUU-------------CCCCU--UUCUGCCCCGCC-CCCAG--UUCAA .....((((..((((((((.......))))))))..(((((....))))).....)))).((((((..(((....-------------.....--.........)))-..)))--))).. ( -29.37) >DroSec_CAF1 69014 111 + 1 UAUUGCACUUAUGCGCUGAUUGCUUUUCAGCGCAAUUUGCUUUUAAGCGAUUCCUAGUGAGGACUGUCGGUUUUU-----CCCCUUUUCCCCU--UUCUGCCCCGCCCCCCAG--CUCAA .....((((..((((((((.......))))))))..(((((....))))).....)))).(..(((..((.....-----.............--...........))..)))--..).. ( -23.75) >DroSim_CAF1 48120 110 + 1 UAUUGCACUUAUGCGCUGAUUGCUUUUCAGCGCAAUUUGCUUUUAAGCGAUUCCUAGUGAGGACUGUCGGUUUUU-----CCGCUUUUCCCCU--UUCUGCCCCGCC-CCCAG--CUCAA .....((((..((((((((.......))))))))..(((((....))))).....)))).(..(((..(((....-----..((.........--....))...)))-..)))--..).. ( -26.92) >DroEre_CAF1 61226 102 + 1 UAUUGCACUUAUGCGCUGAUUGCUUUUCAGCGCAAUUUGCUUUUAAGCGAUUCCUAGUGAGGACUGUCGGUUUUC-------------CCCUU--UUCUGCCACGCC-CCCAG--CUCGA .....((((..((((((((.......))))))))..(((((....))))).....)))).(..(((..(((....-------------.....--.........)))-..)))--..).. ( -25.87) >DroYak_CAF1 62032 104 + 1 UAUUGCACUUAUGCGCUGAUUGCUUUUCAGCGCAAUUUGCUUUUAAGCGAUUCCUAGUGAGGACUGUCGGUUUUC-------------CCCUUUUUUCUGCCCCGCC-CCCUG--CUCGA ....(((....((((((((.......))))))))...)))..(..((((((((((....))))..)))(((....-------------...........))).....-....)--))..) ( -24.06) >DroAna_CAF1 65034 116 + 1 UAUUGCACUUAUGCGCUGAUUGCUUUUCAGCGCAAUUUGCUUUUAAGUGAUUCUUAGUAGAGACCAUCCCUCUACUACCAUCCCUAUUCACCU--GUCUCC-UAGUC-GCCAGGACUCUA ....(((....((((((((.......))))))))...))).....((.(((...((((((((.......))))))))..))).)).....(((--(.(...-.....-).))))...... ( -31.20) >consensus UAUUGCACUUAUGCGCUGAUUGCUUUUCAGCGCAAUUUGCUUUUAAGCGAUUCCUAGUGAGGACUGUCGGUUUUC_____________CCCCU__UUCUGCCCCGCC_CCCAG__CUCAA ....(((....((((((((.......))))))))...))).......((((((((....))))..))))................................................... (-21.65 = -21.27 + -0.39)


| Location | 14,160,920 – 14,161,022 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.19 |
| Mean single sequence MFE | -33.66 |
| Consensus MFE | -21.67 |
| Energy contribution | -21.68 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14160920 102 - 22224390 UUGAA--CUGGG-GGCGGGGCAGAA--AGGGG-------------AAAAACCGACAGUCCUCACUAGGAAUCGCUUAAAAGCAAAUUGCGCUGAAAAGCAAUCAGCGCAUAAGUGCAAUA .....--....(-(((((..(....--.((((-------------(...........))))).....)..))))))....(((...((((((((.......))))))))....))).... ( -30.00) >DroSec_CAF1 69014 111 - 1 UUGAG--CUGGGGGGCGGGGCAGAA--AGGGGAAAAGGGG-----AAAAACCGACAGUCCUCACUAGGAAUCGCUUAAAAGCAAAUUGCGCUGAAAAGCAAUCAGCGCAUAAGUGCAAUA ....(--((...((((((..(....--.(((((...((..-----.....)).....))))).....)..))))))...)))....((((((((.......))))))))........... ( -35.10) >DroSim_CAF1 48120 110 - 1 UUGAG--CUGGG-GGCGGGGCAGAA--AGGGGAAAAGCGG-----AAAAACCGACAGUCCUCACUAGGAAUCGCUUAAAAGCAAAUUGCGCUGAAAAGCAAUCAGCGCAUAAGUGCAAUA ....(--((..(-(((((..(....--.(((((....(((-----.....)))....))))).....)..))))))...)))....((((((((.......))))))))........... ( -37.20) >DroEre_CAF1 61226 102 - 1 UCGAG--CUGGG-GGCGUGGCAGAA--AAGGG-------------GAAAACCGACAGUCCUCACUAGGAAUCGCUUAAAAGCAAAUUGCGCUGAAAAGCAAUCAGCGCAUAAGUGCAAUA ..(((--(((((-(((.(((.....--.....-------------.....)))...))))))).........))))....(((...((((((((.......))))))))....))).... ( -30.06) >DroYak_CAF1 62032 104 - 1 UCGAG--CAGGG-GGCGGGGCAGAAAAAAGGG-------------GAAAACCGACAGUCCUCACUAGGAAUCGCUUAAAAGCAAAUUGCGCUGAAAAGCAAUCAGCGCAUAAGUGCAAUA ..(((--(.(((-(((....(........)((-------------.....))....))))))....(....)))))....(((...((((((((.......))))))))....))).... ( -28.90) >DroAna_CAF1 65034 116 - 1 UAGAGUCCUGGC-GACUA-GGAGAC--AGGUGAAUAGGGAUGGUAGUAGAGGGAUGGUCUCUACUAAGAAUCACUUAAAAGCAAAUUGCGCUGAAAAGCAAUCAGCGCAUAAGUGCAAUA ..((((((((..-.(((.-(....)--.)))...))))(((..(((((((((.....)))))))))...)))))))....(((...((((((((.......))))))))....))).... ( -40.70) >consensus UUGAG__CUGGG_GGCGGGGCAGAA__AGGGG_____________AAAAACCGACAGUCCUCACUAGGAAUCGCUUAAAAGCAAAUUGCGCUGAAAAGCAAUCAGCGCAUAAGUGCAAUA .................((((........((...................))((...(((......))).))))))....(((...((((((((.......))))))))....))).... (-21.67 = -21.68 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:42 2006