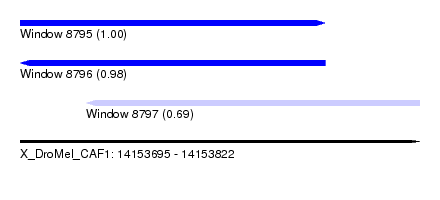
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,153,695 – 14,153,822 |
| Length | 127 |
| Max. P | 0.996929 |

| Location | 14,153,695 – 14,153,792 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 86.06 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -30.65 |
| Energy contribution | -32.90 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14153695 97 + 22224390 C------------UGCUGC-------UGCAUAAAUGCUGAAAGUUUCCUCCAAGGUAGCCUUGAUGUUGCCACUGCACUUUUUGUGCCGCAUGCCGCAGCAAAUUCCCCUUGCCUC .------------((((((-------.(((....(((................((((((......))))))...((((.....)))).)))))).))))))............... ( -28.50) >DroSec_CAF1 61727 104 + 1 C------------UGCUGCGGCGUUUCGCAUAAAUGCAGAAAGUUUCCUCCAAGGUAGCCUUGAUGUUUCCACUGCACUUUUUGUGCCGCAUGCCGCAGCAAAUUCCCCCUGCCUU .------------(((((((((((...((((((((((((...........(((((...))))).((....)))))))...)))))))...)))))))))))............... ( -36.80) >DroSim_CAF1 39482 101 + 1 ---------------CUGCGGCGUUUCGCAUAAAUGCAGAAAGUUUCCUCCAAGGUAGCCUUGAUGUUGCCACUGCACUUUUUGUGCCGCAUGCCGCAGCAAAUUCCCCCUGCCAU ---------------(((((((((...((((((((((((..((....))....((((((......)))))).)))))...)))))))...)))))))))................. ( -34.00) >DroEre_CAF1 53957 114 + 1 CUGCUACUGUUGCUGCUGCGGCGUUUCGCAUAAAUGCAGAAAGUUUCCUCCAAGGUAGCCUUGAUGUUGCCACUGCACUUUUUGUGCCGCAUGCCGCAGCAAAUUCCCCUCGCC-- ..((.......))(((((((((((...((((((((((((..((....))....((((((......)))))).)))))...)))))))...))))))))))).............-- ( -40.20) >consensus C____________UGCUGCGGCGUUUCGCAUAAAUGCAGAAAGUUUCCUCCAAGGUAGCCUUGAUGUUGCCACUGCACUUUUUGUGCCGCAUGCCGCAGCAAAUUCCCCCUGCCUU .............(((((((((((...((((((((((((..((....))....((((((......)))))).)))))...)))))))...)))))))))))............... (-30.65 = -32.90 + 2.25)



| Location | 14,153,695 – 14,153,792 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 86.06 |
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -31.62 |
| Energy contribution | -33.12 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14153695 97 - 22224390 GAGGCAAGGGGAAUUUGCUGCGGCAUGCGGCACAAAAAGUGCAGUGGCAACAUCAAGGCUACCUUGGAGGAAACUUUCAGCAUUUAUGCA-------GCAGCA------------G ...............((((((.(((((..((((.....))))....((.....(((((...)))))(((....)))...))...))))).-------))))))------------. ( -31.40) >DroSec_CAF1 61727 104 - 1 AAGGCAGGGGGAAUUUGCUGCGGCAUGCGGCACAAAAAGUGCAGUGGAAACAUCAAGGCUACCUUGGAGGAAACUUUCUGCAUUUAUGCGAAACGCCGCAGCA------------G ...............(((((((((.....(((....((((((((((....)).(((((...)))))(((....))).)))))))).))).....)))))))))------------. ( -38.80) >DroSim_CAF1 39482 101 - 1 AUGGCAGGGGGAAUUUGCUGCGGCAUGCGGCACAAAAAGUGCAGUGGCAACAUCAAGGCUACCUUGGAGGAAACUUUCUGCAUUUAUGCGAAACGCCGCAG--------------- ..((((((.....))))))(((((.....(((....((((((((((....)).(((((...)))))(((....))).)))))))).))).....)))))..--------------- ( -34.20) >DroEre_CAF1 53957 114 - 1 --GGCGAGGGGAAUUUGCUGCGGCAUGCGGCACAAAAAGUGCAGUGGCAACAUCAAGGCUACCUUGGAGGAAACUUUCUGCAUUUAUGCGAAACGCCGCAGCAGCAACAGUAGCAG --.((....(....((((((((((.....(((....((((((((((....)).(((((...)))))(((....))).)))))))).))).....))))))))))...)....)).. ( -41.10) >consensus AAGGCAAGGGGAAUUUGCUGCGGCAUGCGGCACAAAAAGUGCAGUGGCAACAUCAAGGCUACCUUGGAGGAAACUUUCUGCAUUUAUGCGAAACGCCGCAGCA____________G ...............(((((((((.....(((....((((((((((....)).(((((...)))))(((....))).)))))))).))).....)))))))))............. (-31.62 = -33.12 + 1.50)


| Location | 14,153,716 – 14,153,822 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 87.67 |
| Mean single sequence MFE | -37.43 |
| Consensus MFE | -27.38 |
| Energy contribution | -28.58 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14153716 106 - 22224390 AAGUUGCGGGUU-----GGUUGGGCGUGGUGCGACGAGGCA-AGGGGAAUU--UGCUGCGGCAUGCGGCACAAAAAGUGCAGUGGCAACAUCAAGGCUACCUUGGAGGAAACUU ..(((((.....-----......(((((.(((.....((((-((.....))--))))))).))))).((((.....))))....)))))..(((((...)))))(((....))) ( -35.80) >DroSec_CAF1 61755 106 - 1 AAGUUGCGGGCU-----GGUUGGGCGUGGCGCGGCAAGGCA-GGGGGAAUU--UGCUGCGGCAUGCGGCACAAAAAGUGCAGUGGAAACAUCAAGGCUACCUUGGAGGAAACUU ......((((.(-----((((..(((((.(((((((((...-.......))--))))))).))))).((((.....)))).(((....)))...))))).))))(((....))) ( -40.40) >DroSim_CAF1 39507 106 - 1 AAGUUGCGGGCU-----GGUUGGGCGUGGCGCGGCAUGGCA-GGGGGAAUU--UGCUGCGGCAUGCGGCACAAAAAGUGCAGUGGCAACAUCAAGGCUACCUUGGAGGAAACUU ..(((((.(.((-----......(((((.(((((((.....-.........--))))))).))))).((((.....))))))).)))))..(((((...)))))(((....))) ( -41.84) >DroEre_CAF1 53997 101 - 1 AAGUUGCGGGUU-----GCUUGGACGUGGCGA-----GGCG-AGGGGAAUU--UGCUGCGGCAUGCGGCACAAAAAGUGCAGUGGCAACAUCAAGGCUACCUUGGAGGAAACUU .........(((-----(((..(.((((.((.-----((((-((.....))--)))).)).))))).((((.....))))...))))))..(((((...)))))(((....))) ( -33.80) >DroYak_CAF1 54668 109 - 1 AAGUUGCGAGUUCUGUUGGCUGGCAAGGGCGU-----GGCAAAGGGGAAUUUCUGCUGCGGCAUGCGGCACAAAAAGUGCAGUGGCAACAUCAAGGCUACCUUGGUGGAAACUU ...((((.((((.....)))).))))..((((-----(.(..((.((.....)).))..).))))).((((.....))))(((.....((((((((...))))))))...))). ( -35.30) >consensus AAGUUGCGGGUU_____GGUUGGGCGUGGCGCG_C__GGCA_AGGGGAAUU__UGCUGCGGCAUGCGGCACAAAAAGUGCAGUGGCAACAUCAAGGCUACCUUGGAGGAAACUU ..(((((................(((((.(((.....(((((.((....)).)))))))).))))).((((.....))))....)))))..(((((...)))))(((....))) (-27.38 = -28.58 + 1.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:34 2006