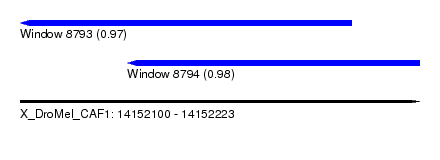
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,152,100 – 14,152,223 |
| Length | 123 |
| Max. P | 0.982894 |

| Location | 14,152,100 – 14,152,202 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.42 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -8.68 |
| Energy contribution | -8.85 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.30 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14152100 102 - 22224390 GC--UGCCUUUCAA-CUUAAGUGAAAUAAAAUACAAUUUAUUUGUAUAUUGUUUUG--CUGUUGACAAUUG--CG----CGACUGCAGAGC-------GCAACUUCUUCGCGAAACGCGA ..--......((((-(...((..((((((.((((((.....)))))).))))))..--)))))))...(((--((----(.........))-------)))).....(((((...))))) ( -29.50) >DroPse_CAF1 71152 87 - 1 GC--UGCUUUUCCAACUUAAGUGAAAUAAAAUACAAUUUAUUUGUAUAUUGUUUUG--CUGUUGACAAUUG--UGCAGGCGGCUGCAG-----------CUCUA---------------- ((--(((.....((((...((..((((((.((((((.....)))))).))))))..--)))))).......--.((.....)).))))-----------)....---------------- ( -24.60) >DroEre_CAF1 52290 102 - 1 GC--UGCCUUUCAA-CUUAAGUGAAAUAAAAUACAAUUUAUUUGUAUAUUGUUUUG--CUGUUGACAAUUG--CG----CGACUACAGAGC-------GCAACUUCUUCGCGAGAGGCGC ((--.(((((((..-(...((..((((((.((((((.....)))))).))))))..--))...)....(((--((----(.........))-------)))).........))))))))) ( -34.00) >DroWil_CAF1 43361 93 - 1 GCUAUGCCUUUCAA-CUUAAGUGAAAUAAAAUACAAUUUAUUUGUAUAUUGUUUUG--CUGUUGACAAUUC--CA----CGACUGCGAAAC-------GG-------UUGCGA----GGC .....(((((((((-(...((..((((((.((((((.....)))))).))))))..--)))))))......--..----((((((.....)-------))-------))).))----))) ( -26.80) >DroYak_CAF1 52698 102 - 1 GC--UGCCUUUCAA-CUUAAGUGAAAUAAAAUACAAUUUAUUUGUAUAUUGUUUUG--CUGUUGACAAUUG--CG----CGACUGCAGAGC-------GCAACUUCUUCGCGCGAGUGGC ((--..(...((((-(...((..((((((.((((((.....)))))).))))))..--)))))))...(((--((----(((.(((.....-------)))......)))))))))..)) ( -33.40) >DroAna_CAF1 57395 97 - 1 GC--UGCCUUUCAA-CUUAAGUGAAAUAAAAUACAAUUUAUUUGUAUAUUGUUUUGUCCUGUUGACAAUUGUUCG----CGACUCUCGUACUGUUGCAGCAACA---------------- ((--(((...((((-(....(..((((((.((((((.....)))))).))))))..)...))))).........(----((.....)))......)))))....---------------- ( -24.90) >consensus GC__UGCCUUUCAA_CUUAAGUGAAAUAAAAUACAAUUUAUUUGUAUAUUGUUUUG__CUGUUGACAAUUG__CG____CGACUGCAGAGC_______GCAACU___UCGCGA_____GC ..........((((.......((((((((.((((((.....)))))).)))))))).....))))....................................................... ( -8.68 = -8.85 + 0.17)

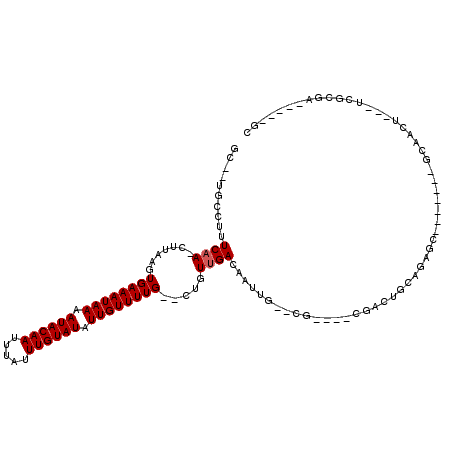
| Location | 14,152,133 – 14,152,223 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.54 |
| Mean single sequence MFE | -26.35 |
| Consensus MFE | -8.68 |
| Energy contribution | -8.85 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.33 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14152133 90 - 22224390 C-----AGCGCGUGG--------------CUCCUUGCGAUGC--UGCCUUUCAA-CUUAAGUGAAAUAAAAUACAAUUUAUUUGUAUAUUGUUUUG--CUGUUGACAAUUG--CG----C .-----.(((((..(--------------(((.....)).))--..)...((((-(...((..((((((.((((((.....)))))).))))))..--))))))).....)--))----) ( -28.00) >DroPse_CAF1 71165 83 - 1 CUCG-----ACGU--------------------------GGC--UGCUUUUCCAACUUAAGUGAAAUAAAAUACAAUUUAUUUGUAUAUUGUUUUG--CUGUUGACAAUUG--UGCAGGC ....-----....--------------------------(.(--(((.....((((...((..((((((.((((((.....)))))).))))))..--))))))((....)--))))).) ( -20.70) >DroEre_CAF1 52323 90 - 1 CUGC-----GCGUGG--------------CUCUUUGCGUUGC--UGCCUUUCAA-CUUAAGUGAAAUAAAAUACAAUUUAUUUGUAUAUUGUUUUG--CUGUUGACAAUUG--CG----C ..((-----(((..(--------------(..........))--..)...((((-(...((..((((((.((((((.....)))))).))))))..--))))))).....)--))----) ( -27.30) >DroWil_CAF1 43383 106 - 1 CUGG-----ACGAGCUUCUUCUUCUUCUACUCCUUGGCGCGCUAUGCCUUUCAA-CUUAAGUGAAAUAAAAUACAAUUUAUUUGUAUAUUGUUUUG--CUGUUGACAAUUC--CA----C .(((-----(.........................((((.....))))..((((-(...((..((((((.((((((.....)))))).))))))..--)))))))....))--))----. ( -23.90) >DroYak_CAF1 52731 95 - 1 CUGCGCAGCGCGUGG--------------CUCCUUGCGAUGC--UGCCUUUCAA-CUUAAGUGAAAUAAAAUACAAUUUAUUUGUAUAUUGUUUUG--CUGUUGACAAUUG--CG----C ..(((((((((((.(--------------(.....)).))))--.))...((((-(...((..((((((.((((((.....)))))).))))))..--)))))))....))--))----) ( -33.80) >DroAna_CAF1 57419 87 - 1 CUGC--AGC--CCGG--------------CU--------GGC--UGCCUUUCAA-CUUAAGUGAAAUAAAAUACAAUUUAUUUGUAUAUUGUUUUGUCCUGUUGACAAUUGUUCG----C ..((--(((--(...--------------..--------)))--)))...((((-(....(..((((((.((((((.....)))))).))))))..)...)))))..........----. ( -24.40) >consensus CUGC_____GCGUGG______________CUCCUUGCG_UGC__UGCCUUUCAA_CUUAAGUGAAAUAAAAUACAAUUUAUUUGUAUAUUGUUUUG__CUGUUGACAAUUG__CG____C ..................................................((((.......((((((((.((((((.....)))))).)))))))).....))))............... ( -8.68 = -8.85 + 0.17)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:31 2006