| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,129,968 – 14,130,059 |
| Length | 91 |
| Max. P | 0.740512 |

| Location | 14,129,968 – 14,130,059 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.17 |
| Mean single sequence MFE | -23.98 |
| Consensus MFE | -14.66 |
| Energy contribution | -14.67 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
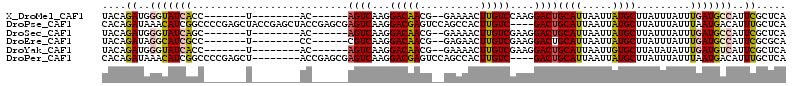
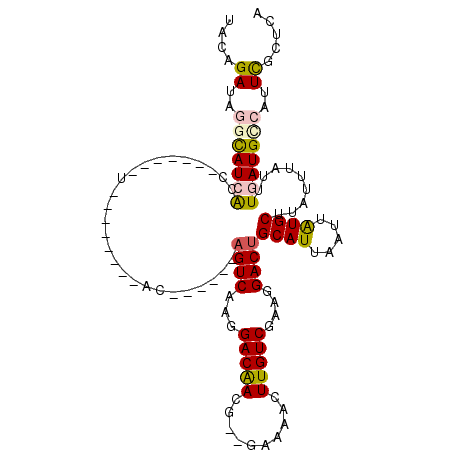
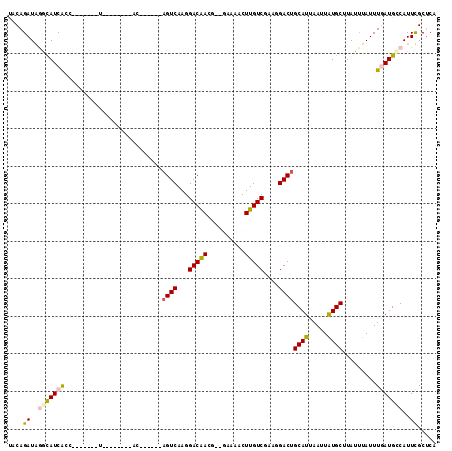
>X_DroMel_CAF1 14129968 91 - 22224390 UACAGAUGGGUAUCACC-------U--------AC------AGUCAAGGACAACG--GAAAACUUGUCCAAGGACUGCAUUAAUUAUGCUUAUUUAUUUGAUGCCAUUCGCUCA ....((..(((((((..-------.--------..------((((..((((((.(--.....)))))))...))))((((.....)))).........)))))))..))..... ( -25.50) >DroPse_CAF1 52562 110 - 1 CACAGAUAAACAUCGGCCCCGAGCUACCGAGCUACCGAGCGAGUCAAGGACGAGUCCAGCCACUUGUC----GACUGCAUUAAUUAUGCUUAUUUAUUUAAUGACAUUUGCUCA ............(((....)))(((....)))....((((((((((..(((((((......)))))))----....((((.....))))............))))..)))))). ( -27.60) >DroSec_CAF1 39636 91 - 1 UACAGAUGGGUAUCAGC-------U--------AC------AGUCAAGGACAACG--GAAAACUUGUCGAAGGACUGCAUUAAUUAUGCUUAUUUAUUUGAUGCCAUUCGCUCA ....((..((((((((.-------.--------..------((((...(((((.(--.....))))))....))))((((.....))))........))))))))..))..... ( -22.40) >DroEre_CAF1 40744 91 - 1 UACAGAUAGGCAUCGCC-------U--------CC------CGUCAAGGACAACG--GAGAACUUGUCGAAGGACUGCAUUAAUUAUGCUUAUUUAUUUGAUGCCAUUCGCGCA ....((..((((((..(-------(--------((------.(((...)))...)--))).....(((....))).((((.....))))..........))))))..))..... ( -24.10) >DroYak_CAF1 38900 91 - 1 UACAGAUGGGUAUCACC-------U--------AC------AGUCAAGGACAACG--GAAAACUUGUCGAAGGACUGCAUUAAUUGUGCUUAUAUAUUUGAUGUCAUUCGCUCA ......(((((......-------.--------.(------((((...(((((.(--.....))))))....)))))((((((.((((....)))).))))))......))))) ( -18.60) >DroPer_CAF1 47847 102 - 1 CACAGAUAAACAUCGGCCCCGAGCU--------ACCGAGCGAGUCAAGGACGAGUCCAGCCACUUGUC----GACUGCAUUAAUUAUGCUUAUUUAUUUAAUGACAUUUGCUCA ............(((....)))...--------...((((((((((..(((((((......)))))))----....((((.....))))............))))..)))))). ( -25.70) >consensus UACAGAUAGGCAUCACC_______U________AC______AGUCAAGGACAACG__GAAAACUUGUCGAAGGACUGCAUUAAUUAUGCUUAUUUAUUUGAUGCCAUUCGCUCA ....((..(((((((..........................((((...(((((..........)))))....))))((((.....)))).........)))))))..))..... (-14.66 = -14.67 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:26 2006