
| Sequence ID | X_DroMel_CAF1 |
|---|---|
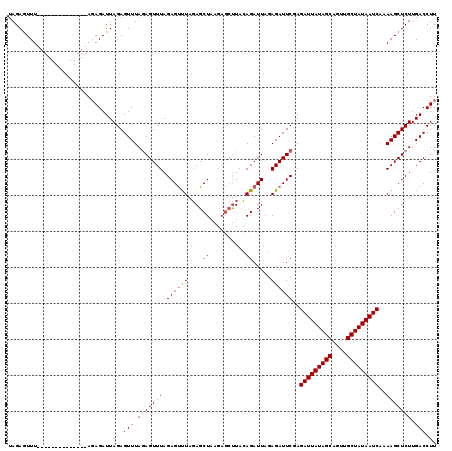
| Location | 14,107,610 – 14,107,748 |
| Length | 138 |
| Max. P | 0.994215 |

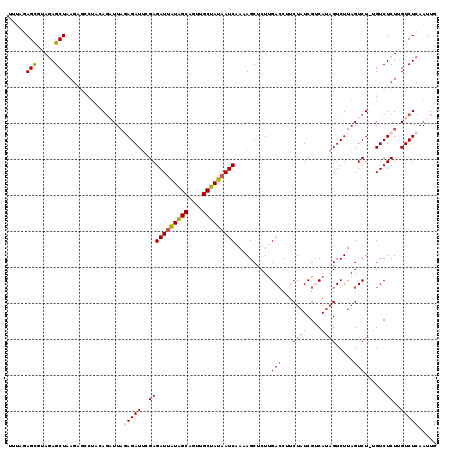
| Location | 14,107,610 – 14,107,709 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.51 |
| Mean single sequence MFE | -32.63 |
| Consensus MFE | -22.02 |
| Energy contribution | -22.47 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.29 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14107610 99 + 22224390 UAGAGUUU--------------ACAGAUUAGAGUUUAGAGUUUAGAGCUUAUAGCU------CUAAAGAUUUGAGAUUCGAGAUUAUAGCAGUUGCUAUAAUCGAAAGCUCUUGACCUU .(((((((--------------.(......((((((.(((((((((((.....)))------)))..))))).))))))..(((((((((....)))))))))).)))))))....... ( -33.10) >DroSec_CAF1 18150 105 + 1 UAGAAUUU--------------AGAGAUUAGAGUUUAGAGUUUAGAGUUUAGAGCUAAGAGCUUACAGAUUAGAGAUUCGAGAUUAUAGCAGUUGCUAUAAUCAAAAGCUCUUGACCUU ........--------------.(((.(((((((((.((((((..(((((.((((.....))))..)))))..))))))..(((((((((....)))))))))..)))))).))).))) ( -29.40) >DroSim_CAF1 18631 119 + 1 UAGAGUUUAGAGUUUAGAGUUUAGAGAUUAGAGUUUAUAGUUUAGAGUCUAGAGCUAAGAGCUUACAGAUUAGAGAUUCGAGAUUAUAGCAGUUGCUAUAAUCAAAAGCUCUUGACCUU .((.(((.(((((((.((((((...((((.((((((.((((((........)))))).))))))...))))..))))))..(((((((((....)))))))))..))))))).))))). ( -35.40) >consensus UAGAGUUU______________AGAGAUUAGAGUUUAGAGUUUAGAGUUUAGAGCUAAGAGCUUACAGAUUAGAGAUUCGAGAUUAUAGCAGUUGCUAUAAUCAAAAGCUCUUGACCUU .............................((.(((.(((((((.((((((....((..........)).....))))))..(((((((((....)))))))))..))))))).))))). (-22.02 = -22.47 + 0.45)


| Location | 14,107,610 – 14,107,709 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.51 |
| Mean single sequence MFE | -23.27 |
| Consensus MFE | -17.52 |
| Energy contribution | -17.63 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -4.42 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14107610 99 - 22224390 AAGGUCAAGAGCUUUCGAUUAUAGCAACUGCUAUAAUCUCGAAUCUCAAAUCUUUAG------AGCUAUAAGCUCUAAACUCUAAACUCUAAUCUGU--------------AAACUCUA .((((..((((.....(((((((((....)))))))))..............(((((------(((.....))))))))))))..)).)).......--------------........ ( -25.50) >DroSec_CAF1 18150 105 - 1 AAGGUCAAGAGCUUUUGAUUAUAGCAACUGCUAUAAUCUCGAAUCUCUAAUCUGUAAGCUCUUAGCUCUAAACUCUAAACUCUAAACUCUAAUCUCU--------------AAAUUCUA .(((..((((((((..(((((((((....)))))))))..((........))...))))))))..).))............................--------------........ ( -21.80) >DroSim_CAF1 18631 119 - 1 AAGGUCAAGAGCUUUUGAUUAUAGCAACUGCUAUAAUCUCGAAUCUCUAAUCUGUAAGCUCUUAGCUCUAGACUCUAAACUAUAAACUCUAAUCUCUAAACUCUAAACUCUAAACUCUA .(((((.((((((...(((((((((....)))))))))..((........))...........)))))).))).))........................................... ( -22.50) >consensus AAGGUCAAGAGCUUUUGAUUAUAGCAACUGCUAUAAUCUCGAAUCUCUAAUCUGUAAGCUCUUAGCUCUAAACUCUAAACUCUAAACUCUAAUCUCU______________AAACUCUA .((((..((((((...(((((((((....)))))))))..((........))...........))))))..)).))........................................... (-17.52 = -17.63 + 0.11)

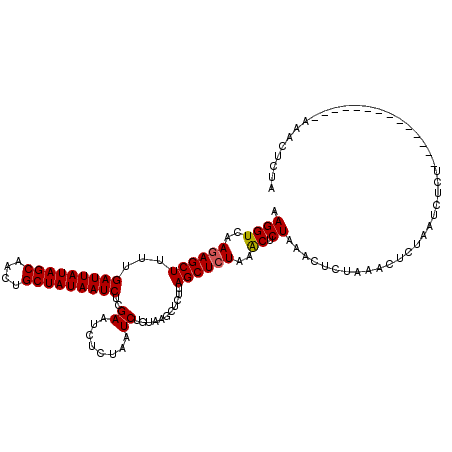

| Location | 14,107,636 – 14,107,748 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.46 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -17.16 |
| Energy contribution | -17.80 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14107636 112 + 22224390 UUUAGAGCUUAUAGCU------CUAAAGAUUUGAGAUUCGAGAUUAUAGCAGUUGCUAUAAUCGAAAGCUCUUGACCUUCUAUCGUCAUAGUCUUAGUCU-UGUCUCUUGUCUCAAUUG ((((((((.....)))------)))))(((..(((((..(((((((((((....)))))))))..(((((..((((........)))).)).)))..)).-.)))))..)))....... ( -35.90) >DroSec_CAF1 18176 118 + 1 UUUAGAGUUUAGAGCUAAGAGCUUACAGAUUAGAGAUUCGAGAUUAUAGCAGUUGCUAUAAUCAAAAGCUCUUGACCUUCUAUCGUCAUAGUCUUAGUCU-UGUCUCUUGUCUCAAUUG ....(((...((((.(((((.((...(((((((((......(((((((((....))))))))).....))).((((........)))))))))).)))))-)).))))...)))..... ( -33.90) >DroSim_CAF1 18671 118 + 1 UUUAGAGUCUAGAGCUAAGAGCUUACAGAUUAGAGAUUCGAGAUUAUAGCAGUUGCUAUAAUCAAAAGCUCUUGACCUUCUAUCGUCAUAGUCUUAGUCU-UGUCUCUUGUCUCAAUUG ....(((.(.((((.(((((.((...(((((((((......(((((((((....))))))))).....))).((((........)))))))))).)))))-)).)))).).)))..... ( -34.80) >DroEre_CAF1 19572 112 + 1 -----AGCGUAGAGCUUAGAGCCUAGACCUUAUAGAUUAGAGAUGGUAGCAGUUGCUAUGAUCUA-AGCCCUCGACCAUCUAUCGCCAUAGUCUUAGUCU-UGUCUCUUGUCUCAAUUG -----......(((...((((.(.((((.....((((((.(((((((.(.((..(((........-))).))).))))))).......))))))..))))-.).))))...)))..... ( -26.80) >DroYak_CAF1 17809 108 + 1 -----AGCGUAGAGCUUAGU-----GAGCUUAUAGAUAAGAGAUUAUGGCAAUUGCUAUAAUCGA-AGCCCUUGAACAUCUUUCGACACCGUCUUAGUCUUUGUCUCUCGUCUCAAUUG -----.(((.(((((..((.-----..(((...((((....(((((((((....)))))))))..-.....(((((.....)))))....)))).)))))..).)))))))........ ( -23.50) >consensus UUUAGAGCGUAGAGCUAAGAGCCUACAGAUUAGAGAUUCGAGAUUAUAGCAGUUGCUAUAAUCAAAAGCUCUUGACCUUCUAUCGUCAUAGUCUUAGUCU_UGUCUCUUGUCUCAAUUG .....(((.....)))................(((((..(((((((((((....)))))))))..........(((...((((....)))).....)))......))..)))))..... (-17.16 = -17.80 + 0.64)


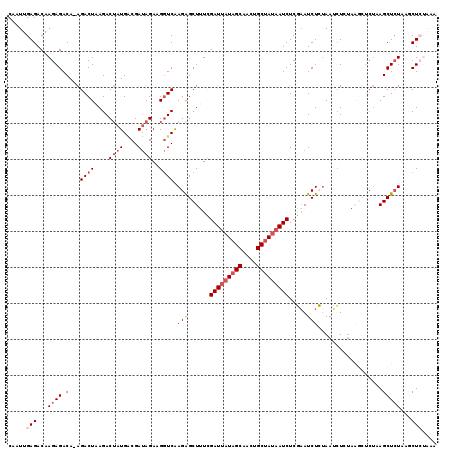
| Location | 14,107,636 – 14,107,748 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.46 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -16.36 |
| Energy contribution | -17.92 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14107636 112 - 22224390 CAAUUGAGACAAGAGACA-AGACUAAGACUAUGACGAUAGAAGGUCAAGAGCUUUCGAUUAUAGCAACUGCUAUAAUCUCGAAUCUCAAAUCUUUAG------AGCUAUAAGCUCUAAA ...((((((...((....-....................((((((.....))))))(((((((((....)))))))))))...))))))....((((------(((.....))))))). ( -32.20) >DroSec_CAF1 18176 118 - 1 CAAUUGAGACAAGAGACA-AGACUAAGACUAUGACGAUAGAAGGUCAAGAGCUUUUGAUUAUAGCAACUGCUAUAAUCUCGAAUCUCUAAUCUGUAAGCUCUUAGCUCUAAACUCUAAA ...........((((...-(((((((((((.((((........))))((((.(((.(((((((((....)))))))))..))).))))........)).)))))).)))...))))... ( -31.30) >DroSim_CAF1 18671 118 - 1 CAAUUGAGACAAGAGACA-AGACUAAGACUAUGACGAUAGAAGGUCAAGAGCUUUUGAUUAUAGCAACUGCUAUAAUCUCGAAUCUCUAAUCUGUAAGCUCUUAGCUCUAGACUCUAAA ...........((((.(.-(((((((((((.((((........))))((((.(((.(((((((((....)))))))))..))).))))........)).)))))).))).).))))... ( -32.10) >DroEre_CAF1 19572 112 - 1 CAAUUGAGACAAGAGACA-AGACUAAGACUAUGGCGAUAGAUGGUCGAGGGCU-UAGAUCAUAGCAACUGCUACCAUCUCUAAUCUAUAAGGUCUAGGCUCUAAGCUCUACGCU----- ...........((((.(.-.......((((((........)))))).((((((-((((((.((((....)))).......((.....)).))))))))))))..))))).....----- ( -27.90) >DroYak_CAF1 17809 108 - 1 CAAUUGAGACGAGAGACAAAGACUAAGACGGUGUCGAAAGAUGUUCAAGGGCU-UCGAUUAUAGCAAUUGCCAUAAUCUCUUAUCUAUAAGCUC-----ACUAAGCUCUACGCU----- ......((.((((((.(.........((..(..((....))..)))..(((((-(.((((((.((....)).))))))..........))))))-----.....))))).))))----- ( -21.60) >consensus CAAUUGAGACAAGAGACA_AGACUAAGACUAUGACGAUAGAAGGUCAAGAGCUUUCGAUUAUAGCAACUGCUAUAAUCUCGAAUCUCUAAUCUCUAAGCUCUAAGCUCUAAGCUCUAAA .....(((...((((.(...((((....((((....))))..)))).(((......(((((((((....))))))))).....)))..................)))))...))).... (-16.36 = -17.92 + 1.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:22 2006