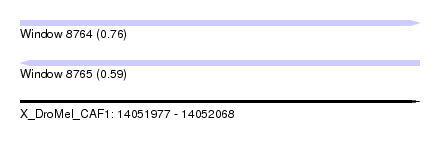
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,051,977 – 14,052,068 |
| Length | 91 |
| Max. P | 0.763244 |

| Location | 14,051,977 – 14,052,068 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 82.72 |
| Mean single sequence MFE | -17.70 |
| Consensus MFE | -12.60 |
| Energy contribution | -13.35 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
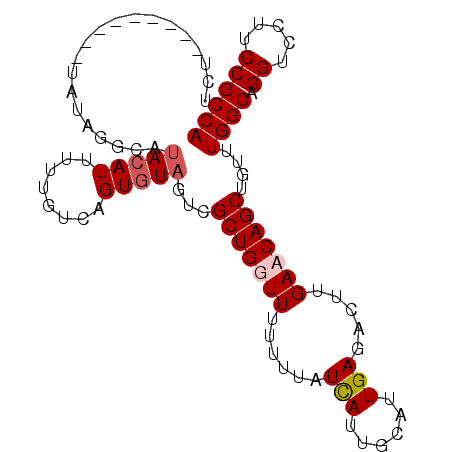
>X_DroMel_CAF1 14051977 91 + 22224390 UGGCGGAAGGACCUGCCAAACAGCUGUUCAAGUCUCAAUGCAAUGAUAAAAAAAACAGCGACUACACUGACAAAUAUGUGUGCACAUA---------AAA ((((((......))))))....((((((......(((......))).......))))))...(((((..........)))))......---------... ( -19.12) >DroSec_CAF1 20397 91 + 1 UGGCGGAAGGACCUGCCAAACAGCUGUUCAAGUCUCAAUGCAAUGAUAAAAAAAACAGCGACUACACUGAAAAAAAUGUAAGCGUGUA---------AAA ((((((......))))))....((((((......(((......))).......))))))...(((((................)))))---------... ( -18.61) >DroEre_CAF1 20782 100 + 1 UGGCGGAAGGACCUGCCAAACAGCUGUUCCAGUCUCAAUGCAAUAAUAAAAAAACCAGCAGCUACACAGACAAAAAUGUAUAGCUAUAUAGAUAAAAAGA ((((((......))))))....((((.............................))))(((((.(((........))).)))))............... ( -19.05) >DroYak_CAF1 20292 91 + 1 UGGCGGAAGGACCUGCCAAACAGCUGUUCAAGUCUCAAUACAAUGACAAAUAAACCAGCAACUACACUGACACAAAUUUAAAACUAUA---------AGA ((((((......))))))....((((.....(((..........)))........)))).............................---------... ( -14.02) >consensus UGGCGGAAGGACCUGCCAAACAGCUGUUCAAGUCUCAAUGCAAUGAUAAAAAAAACAGCAACUACACUGACAAAAAUGUAAACCUAUA_________AAA ((((((......))))))....((((((......(((......))).......))))))......................................... (-12.60 = -13.35 + 0.75)



| Location | 14,051,977 – 14,052,068 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.72 |
| Mean single sequence MFE | -21.25 |
| Consensus MFE | -14.69 |
| Energy contribution | -15.50 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14051977 91 - 22224390 UUU---------UAUGUGCACACAUAUUUGUCAGUGUAGUCGCUGUUUUUUUUAUCAUUGCAUUGAGACUUGAACAGCUGUUUGGCAGGUCCUUCCGCCA ...---------....((((((((....)))..)))))...(((((((......(((......))).....)))))))....((((.((.....)))))) ( -22.10) >DroSec_CAF1 20397 91 - 1 UUU---------UACACGCUUACAUUUUUUUCAGUGUAGUCGCUGUUUUUUUUAUCAUUGCAUUGAGACUUGAACAGCUGUUUGGCAGGUCCUUCCGCCA ..(---------(((((................))))))..(((((((......(((......))).....)))))))....((((.((.....)))))) ( -21.19) >DroEre_CAF1 20782 100 - 1 UCUUUUUAUCUAUAUAGCUAUACAUUUUUGUCUGUGUAGCUGCUGGUUUUUUUAUUAUUGCAUUGAGACUGGAACAGCUGUUUGGCAGGUCCUUCCGCCA ...............(((((((((........)))))))))((((.((((....(((......)))....))))))))....((((.((.....)))))) ( -23.00) >DroYak_CAF1 20292 91 - 1 UCU---------UAUAGUUUUAAAUUUGUGUCAGUGUAGUUGCUGGUUUAUUUGUCAUUGUAUUGAGACUUGAACAGCUGUUUGGCAGGUCCUUCCGCCA ...---------.((((((............((((......))))(((((..(.(((......))).)..))))))))))).((((.((.....)))))) ( -18.70) >consensus UCU_________UAUAGGCAUACAUUUUUGUCAGUGUAGUCGCUGGUUUUUUUAUCAUUGCAUUGAGACUUGAACAGCUGUUUGGCAGGUCCUUCCGCCA ....................(((((........)))))...(((((((......(((......))).....)))))))....((((.((.....)))))) (-14.69 = -15.50 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:05 2006