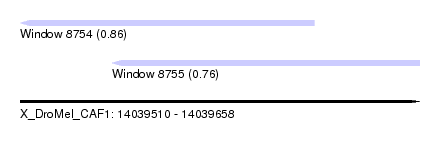
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,039,510 – 14,039,658 |
| Length | 148 |
| Max. P | 0.855630 |

| Location | 14,039,510 – 14,039,619 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 87.95 |
| Mean single sequence MFE | -41.62 |
| Consensus MFE | -28.70 |
| Energy contribution | -27.78 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14039510 109 - 22224390 UUUCCGGCUUCAAGGACCCAAGGACAUUGAGUUCGCCUGCCUAUCGUGGUGGUCUCAAUGUCUGGGUAAAUUGUACUGGAUUAUCUG------GCGAGUGAAAUGCGAGAUUGGC ..(((((...(((..(((((..(((((((((..((((..........))))..))))))))))))))...)))..)))))..((((.------(((.......))).)))).... ( -40.60) >DroSec_CAF1 8618 109 - 1 UUUCCGGCAUCAAGGACCCAAGGACAUUGAGUUCGCCUGCCUGUCGGGCUGGUCUCAAUGUCUGGGUAAAUUGUACUGGACUAUCUG------GCGAGUGAAAUGCGAGAUUGGC ..(((((...(((..(((((..(((((((((...(((.(((.....))).)))))))))))))))))...)))..)))))(.((((.------(((.......))).)))).).. ( -42.80) >DroSim_CAF1 7518 109 - 1 UUUCCGGCAUCAAGGACCCAAGGACAUUGAGUUCGCCUGCCUGUCGGGUUGCUCUCAAUGCCUGGGUAAAUUGUACUGGACUAUCUG------GCGAGUGAAAUGCGAGAUUGGC ..(((((...(((..(((((.((.(((((((..((((((.....)))).))..))))))))))))))...)))..)))))(.((((.------(((.......))).)))).).. ( -39.60) >DroEre_CAF1 8426 106 - 1 UUUCCGGCAUCAAGGGCCCAAGAACGUUGGGUCUGCCCGCAUGUCGG--UGGGCUCAAUGUCUGGGUAAAUUG-ACUGGAUUAUCUG------GCGAGUGAAAUGCGAGAUUGGC ..(((((..((((..(((((.((.(((((((((..((........))--..))))))))))))))))...)))-))))))..((((.------(((.......))).)))).... ( -41.90) >DroYak_CAF1 8257 113 - 1 UUUCCGGCUUCAAGGGCCCAGGAACGUUGAGUUUGCCAGCAUGUUGG--UGGGCUCAAUGUCUGGGUAAAUUGUACUGGAUUAUCUGUGAAAUGCGAGUGAAAUGCGAGAUGGGC ..(((((...(((..((((((..((((((((((..((((....))))--..))))))))))))))))...)))..)))))((((((((.....))..((.....)).)))))).. ( -43.20) >consensus UUUCCGGCAUCAAGGACCCAAGGACAUUGAGUUCGCCUGCCUGUCGGG_UGGUCUCAAUGUCUGGGUAAAUUGUACUGGAUUAUCUG______GCGAGUGAAAUGCGAGAUUGGC ..(((((...(((..((((.(((.(((((((..((((((.....)))).))..))))))))))))))...)))..))))).............(((.......)))......... (-28.70 = -27.78 + -0.92)


| Location | 14,039,544 – 14,039,658 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 87.02 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -23.16 |
| Energy contribution | -22.64 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14039544 114 - 22224390 UCUUGAUUUUCUCUCUCUCUCAAU-CAAUUGCAGUUAGAUUUUCCGGCUUCAAGGACCCAAGGACAUUGAGUUCGCCUGCCUAUCGUGGUGGUCUCAAUGUCUGGGUAAAUUGUA ((((((.......(((..(((((.-...))).))..)))..........))))))(((((..(((((((((..((((..........))))..))))))))))))))........ ( -32.83) >DroSec_CAF1 8652 114 - 1 UCUUGAUUUUCUCUAUCUCUCAAU-CAAUUGCAGUUAGAUUUUCCGGCAUCAAGGACCCAAGGACAUUGAGUUCGCCUGCCUGUCGGGCUGGUCUCAAUGUCUGGGUAAAUUGUA (((((((.......(((((((((.-...))).))..))))........)))))))(((((..(((((((((...(((.(((.....))).)))))))))))))))))........ ( -35.06) >DroSim_CAF1 7552 114 - 1 UCUUGAUUUUCUCUAUCUCUCAAU-CAAUUGCAGUUAGAUUUUCCGGCAUCAAGGACCCAAGGACAUUGAGUUCGCCUGCCUGUCGGGUUGCUCUCAAUGCCUGGGUAAAUUGUA (((((((.......(((((((((.-...))).))..))))........)))))))(((((.((.(((((((..((((((.....)))).))..))))))))))))))........ ( -31.86) >DroEre_CAF1 8460 109 - 1 UCUUGAUCCUCCC--UCUAUCAAU-CAAUUUCAGUUAGAUUUUCCGGCAUCAAGGGCCCAAGAACGUUGGGUCUGCCCGCAUGUCGG--UGGGCUCAAUGUCUGGGUAAAUUG-A (((((((((....--((((.....-..........))))......)).)))))))(((((.((.(((((((((..((........))--..))))))))))))))))......-. ( -35.36) >DroYak_CAF1 8297 111 - 1 UCUUGAUUUUCUC--CCUCUCAACCCAAUUUCAGUUAGAUUUUCCGGCUUCAAGGGCCCAGGAACGUUGAGUUUGCCAGCAUGUUGG--UGGGCUCAAUGUCUGGGUAAAUUGUA ((((((.......--..(((.(((.........))))))..........))))))((((((..((((((((((..((((....))))--..))))))))))))))))........ ( -36.51) >consensus UCUUGAUUUUCUCU_UCUCUCAAU_CAAUUGCAGUUAGAUUUUCCGGCAUCAAGGACCCAAGGACAUUGAGUUCGCCUGCCUGUCGGG_UGGUCUCAAUGUCUGGGUAAAUUGUA .............................(((((((.......((........))((((.(((.(((((((..((((((.....)))).))..)))))))))))))).))))))) (-23.16 = -22.64 + -0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:56 2006