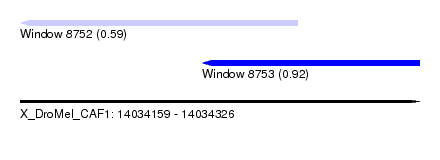
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,034,159 – 14,034,326 |
| Length | 167 |
| Max. P | 0.919515 |

| Location | 14,034,159 – 14,034,275 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.43 |
| Mean single sequence MFE | -34.33 |
| Consensus MFE | -21.58 |
| Energy contribution | -22.08 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
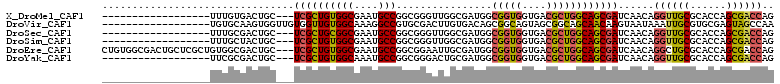
>X_DroMel_CAF1 14034159 116 - 22224390 GACGCUGGCAGCGAUCAACAGGUUGCGCACCAGCGACCAGCGAAGGGGCAAGCAACUGGCGGAGAUGCGAUAUUGCAUUCAGAUGCACUUUGCACGGUGAAAAGCAAAACAAUAAU---- ..((((((..((((((....))))))...))))))(((.((((((..(((.((.....))...((((((....))))))....))).))))))..)))..................---- ( -37.70) >DroVir_CAF1 21403 111 - 1 AGCGGCAGCAACAAGUAAUAAAUUGCGUGCGAGUAGCCAACGAGAGGGCACCGACCUUGCGUUUCUCAGAUAUUGCAUU-AAAUGCACUUUGCUCGGCCAAUC---AAACGUU-GC---- .(((((.(((....((((....)))).)))((...(((...(((((((((.......))).))))))......(((((.-..)))))........)))...))---....)))-))---- ( -27.80) >DroSec_CAF1 3199 116 - 1 GACGCUGGCAGCGAUCAACAGGUUGCGCACCAGCGACCAGCGAAAGGGCAAGCAACUGGCAGAUAUGCGAUAUUGCAUUCAGAUGCACUUUGCACGGCUGAAAACAAAACAAUAAU---- ..((((((..((((((....))))))...)))))).(((((....).(....)..))))(((...(((((...(((((....)))))..)))))...)))................---- ( -35.60) >DroSim_CAF1 921 116 - 1 GACGCUGGCAGCGAUCAACAGGUUGCGCACCAGCGACCAGCGAAAGGGCAAGCAACUGGCGGAGAUGCGAUAUUGCAUUCAUAUGCACUUUGCACGGCUGAAAACAAAACAAUAAU---- ..(((((((.((((((....))))))((....))).))))))....(((..((((...(((..((((((....))))))....)))...))))...))).................---- ( -35.20) >DroEre_CAF1 3112 120 - 1 GACGCUGGCAGCGAUCAACAGGCUGCGCACCAGCGACCAGCGAAAGGGCAAGAACCUUGCACAGAUCCGAUAUUGCAUUCAGAUGCACUUUGCACGGCGGAAAUCAAGACAAUAAAUACG ..(((((((((.((((.....(((((((....)))..))))......(((((...)))))...))))......(((((....)))))..)))).)))))..................... ( -33.10) >DroYak_CAF1 3099 118 - 1 GACGCUGGCAGCGAUCAACAGGUUGCGCACCAGCGACCAGCGAAAGGGCAAGAACCUUGCACAGAUCCGAUAUUGCAUUCAGAUGCACUUUGCACAGCGGAAAACAAGACAAUAAAUA-- ..(((((((((.((((....((((((......))))))..(....).(((((...)))))...))))......(((((....)))))..)))).)))))...................-- ( -36.60) >consensus GACGCUGGCAGCGAUCAACAGGUUGCGCACCAGCGACCAGCGAAAGGGCAAGCAACUGGCAGAGAUCCGAUAUUGCAUUCAGAUGCACUUUGCACGGCGGAAAACAAAACAAUAAU____ ..((((((..((((((....))))))...))))))....((.((((.((.........)).............(((((....)))))))))))........................... (-21.58 = -22.08 + 0.50)

| Location | 14,034,235 – 14,034,326 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.26 |
| Mean single sequence MFE | -42.20 |
| Consensus MFE | -28.51 |
| Energy contribution | -28.60 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
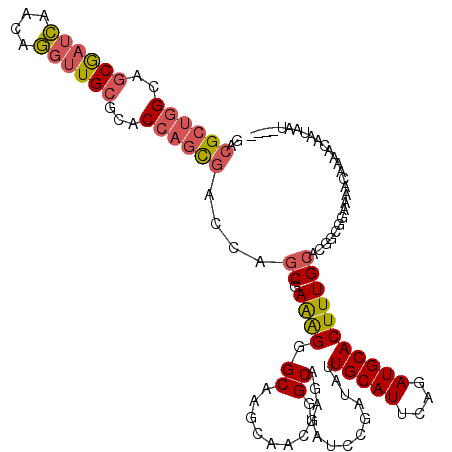
>X_DroMel_CAF1 14034235 91 - 22224390 ------------------UUUGUGACUGC---UCGCUGUGGCGAAUGCCGGCGGGUUGGCGAUGGCGGUGGUGACGCUGGCAGCGAUCAACAGGUUGCGCACCAGCGACCAG ------------------.......(((.---((((((..(((...(((.....(((((((.((.(((((....))))).)).)).))))).)))..)))..)))))).))) ( -42.50) >DroVir_CAF1 21474 94 - 1 ------------------UGUGCAAGUGGUUGUGGUUGUGGCAAAGGCCGUGCGACUUGUGACAGCGGCAGUAGCGGCAGCAACAAGUAAUAAAUUGCGUGCGAGUAGCCAA ------------------...((((.((.((((.(((((.((.(..(((((.((.....))...)))))..).)).))))).)))).)).....))))(.((.....))).. ( -29.60) >DroSec_CAF1 3275 91 - 1 ------------------UUUGCGACUGC---UCGCUGCGGCGAAUGCCGGCGGGUUGGCGAUGGCGGUGGUGACGCUGGCAGCGAUCAACAGGUUGCGCACCAGCGACCAG ------------------.......(((.---((((((..(((...(((.....(((((((.((.(((((....))))).)).)).))))).)))..)))..)))))).))) ( -43.30) >DroSim_CAF1 997 91 - 1 ------------------UUUGCUACUGC---UCGCUGUGGCGAAUGCCGGCGGGUUGGCGAUGGCGGUGGUGACGCUGGCAGCGAUCAACAGGUUGCGCACCAGCGACCAG ------------------.(..(((((((---((((....)))).((((((....))))))...)))))))..)((((((..((((((....))))))...))))))..... ( -42.80) >DroEre_CAF1 3192 109 - 1 CUGUGGCGACUGCUCGCUGUGGCGACUGC---UCGCUGUGGCGAAUGCCGGCGGAAUUGCGAUGGCGGUGGUGACGCUGGCAGCGAUCAACAGGCUGCGCACCAGCGACCAG (((..((((....))))..)))...(((.---((((((..(((...(((.(..((.((((..((.(((((....))))).))))))))..).)))..)))..)))))).))) ( -50.10) >DroYak_CAF1 3177 91 - 1 ------------------UUCGCGACUGC---UCGCUGUGGCAAAUGCCGGCGGGACUGCGAUGGCGGUGGUGACGCUGGCAGCGAUCAACAGGUUGCGCACCAGCGACCAG ------------------.......(((.---((((((..((...((((((((..((..(.......)..))..))))))))((((((....))))))))..)))))).))) ( -44.90) >consensus __________________UUUGCGACUGC___UCGCUGUGGCGAAUGCCGGCGGGUUGGCGAUGGCGGUGGUGACGCUGGCAGCGAUCAACAGGUUGCGCACCAGCGACCAG ................................((((((((((....)))................(((((....))))))))))))......((((((......)))))).. (-28.51 = -28.60 + 0.09)
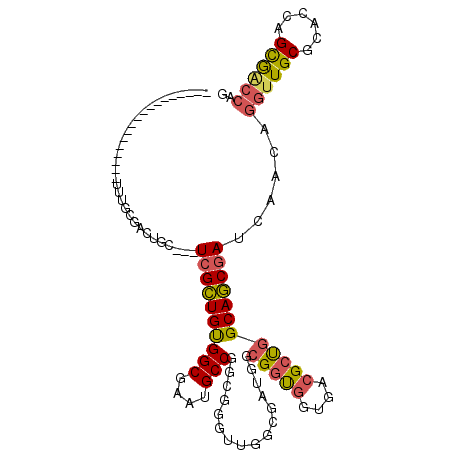
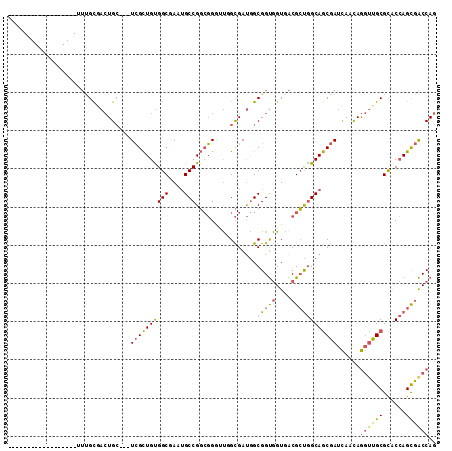
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:54 2006