| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,027,733 – 14,027,840 |
| Length | 107 |
| Max. P | 0.799842 |

| Location | 14,027,733 – 14,027,840 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.83 |
| Mean single sequence MFE | -32.91 |
| Consensus MFE | -22.40 |
| Energy contribution | -22.97 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14027733 107 + 22224390 UUA------AAUGGAGAGGGCAUAGCUACUAUGGAGUCGGAUUAACCAGAUUUCUUUAGCUAUAAUUUUGAUGGUUAGGGAAUCCUAUCUAGAGAUGCCACAUCCCCAUAUUC------- ...------.((((.((.(((((..(((....((((((((.....)).)))((((((((((((.......)))))))))))))))....)))..)))))...)).))))....------- ( -32.10) >DroSec_CAF1 54366 120 + 1 UUAAUCCAAAAUGGAGAGGGCAUAGCUCCUAUGGAAUUGGGUUAACCCGAUUUCUUUAGACAUCAUUUUGGUGGUUAGGGAAUCCUAUCUAGAGAUACUACAUCCCCAUAUUCCUAGCAA .....(((((((((...((((...))))(((.(((((((((....)))))))))..)))...)))))))))..(((((((((((((....)).)))..............)))))))).. ( -33.03) >DroSim_CAF1 54137 120 + 1 UUAAUCCAAAAUGGAGAGGGCAUAGCUCCUAUGGAAUCGGGUUAACCCGAUUUCUUUAGAUAUCAUUUUGAUGGUAAGGGAAUCCUAUCUAGAGAUACUACAUCCCCAUAUUCCUGGCAA .....(((....((((.((((...)))).((((((((((((....)))))))((((((((((((((....))))))(((....))).))))))))..........))))))))))))... ( -33.60) >consensus UUAAUCCAAAAUGGAGAGGGCAUAGCUCCUAUGGAAUCGGGUUAACCCGAUUUCUUUAGAUAUCAUUUUGAUGGUUAGGGAAUCCUAUCUAGAGAUACUACAUCCCCAUAUUCCU_GCAA ..........((((.((.(((((((...))))).(((((((....)))))))((((((((((((.....))))..((((....))))))))))))..))...)).))))........... (-22.40 = -22.97 + 0.57)

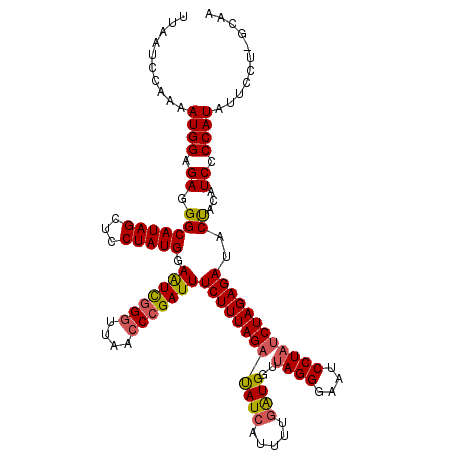

| Location | 14,027,733 – 14,027,840 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.83 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -22.55 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14027733 107 - 22224390 -------GAAUAUGGGGAUGUGGCAUCUCUAGAUAGGAUUCCCUAACCAUCAAAAUUAUAGCUAAAGAAAUCUGGUUAAUCCGACUCCAUAGUAGCUAUGCCCUCUCCAUU------UAA -------....(((((((.(.(((.........((((....))))............(((((((........(((.....))).........)))))))))))))))))).------... ( -25.53) >DroSec_CAF1 54366 120 - 1 UUGCUAGGAAUAUGGGGAUGUAGUAUCUCUAGAUAGGAUUCCCUAACCACCAAAAUGAUGUCUAAAGAAAUCGGGUUAACCCAAUUCCAUAGGAGCUAUGCCCUCUCCAUUUUGGAUUAA ......((....((((((.((..((((....))))..)))))))).)).((((((((...............(((....)))........(((.((...)))))...))))))))..... ( -28.40) >DroSim_CAF1 54137 120 - 1 UUGCCAGGAAUAUGGGGAUGUAGUAUCUCUAGAUAGGAUUCCCUUACCAUCAAAAUGAUAUCUAAAGAAAUCGGGUUAACCCGAUUCCAUAGGAGCUAUGCCCUCUCCAUUUUGGAUUAA ...((((((....((((..(((((.(((.((((((((....))....(((....))).)))))).)))(((((((....)))))))........)))))..))))....))))))..... ( -33.60) >consensus UUGC_AGGAAUAUGGGGAUGUAGUAUCUCUAGAUAGGAUUCCCUAACCAUCAAAAUGAUAUCUAAAGAAAUCGGGUUAACCCGAUUCCAUAGGAGCUAUGCCCUCUCCAUUUUGGAUUAA ...((((((....((((..(((((.........((((....))))...............((((..(((.(((((....))))))))..)))).)))))..))))....))))))..... (-22.55 = -23.00 + 0.45)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:51 2006