| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,546,165 – 1,546,258 |
| Length | 93 |
| Max. P | 0.976502 |

| Location | 1,546,165 – 1,546,258 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 82.97 |
| Mean single sequence MFE | -34.37 |
| Consensus MFE | -28.38 |
| Energy contribution | -29.96 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.04 |
| Mean z-score | -4.40 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
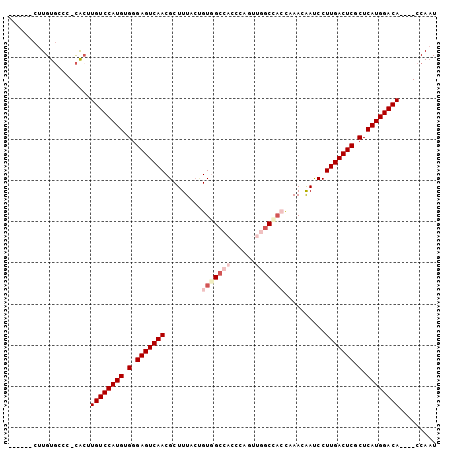
>X_DroMel_CAF1 1546165 93 + 22224390 ------CUUGUGCCA-CAAUUGUCCAUGUGGGAGUCAACGCUUUACUGUUGCCACCCAUUUGGCCACCAAACAAUCCUUGACUCGCUCAUGGACA----CCAAU ------.........-....((((((((.(.(((((((.........((.((((......)))).))..........))))))).).))))))))----..... ( -29.91) >DroSec_CAF1 8546 93 + 1 ------CUUGUGCUC-CACUUGUCCAUGUGGGAGUCAACGCUUUACUGUGGCCACCCAUUUGGCCACCAAACAACCCUUGACUCGCUCAUGGACA----CCAAU ------.........-....((((((((.(.(((((((.........(((((((......)))))))..........))))))).).))))))))----..... ( -36.41) >DroSim_CAF1 9565 93 + 1 ------CUUGUGCUC-CACUUGUCCAUGUGGGAGUCAACGCUUUACUGUAGCCACCCAUUUGGCCACCAAACAACCCUUGACUCGCUCAUGGACA----CCAAU ------.........-....((((((((.(.(((((((.........((.((((......)))).))..........))))))).).))))))))----..... ( -29.91) >DroEre_CAF1 10103 93 + 1 ------CUUGUGUCC-CACUUGUCCAUGUGGGAGUCAACGCUUUACUGUGGCCACCCAGUUGGCCAAUCAACAAUCCUUGACUCGCUCAUGGACA----CCAAU ------.........-....((((((((.(.(((((((..........((((((......))))))...........))))))).).))))))))----..... ( -34.50) >DroYak_CAF1 8749 90 + 1 ------CUUGUGUCC-CACUUGUCCAUGUGGGAGUCAACGCUUUACCGUGGCCACCCAG---GCCAAUCAACAAUCCUUGACUCGCUCAUGGACA----CCAAU ------.........-....((((((((.(.(((((((..........(((((.....)---))))...........))))))).).))))))))----..... ( -33.50) >DroPer_CAF1 12688 104 + 1 AUUCCGCUUGUCCGCGUGCAUGUCCAUGUGGGAGUCAACGCUUUAGAGUGGAGGAAGUGGAGGCUCCCCAACGAUCCUUGACUCGCUCAUGGACAUGACCCACU ....(((......)))..((((((((((.(.(((((..((((....))))(((((..(((.(....))))....)))))))))).).))))))))))....... ( -42.00) >consensus ______CUUGUGCCC_CACUUGUCCAUGUGGGAGUCAACGCUUUACUGUGGCCACCCAGUUGGCCACCAAACAAUCCUUGACUCGCUCAUGGACA____CCAAU ....................((((((((.(.(((((((.........(((((((......)))))))..........))))))).).))))))))......... (-28.38 = -29.96 + 1.58)


| Location | 1,546,165 – 1,546,258 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 82.97 |
| Mean single sequence MFE | -36.62 |
| Consensus MFE | -29.87 |
| Energy contribution | -31.62 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
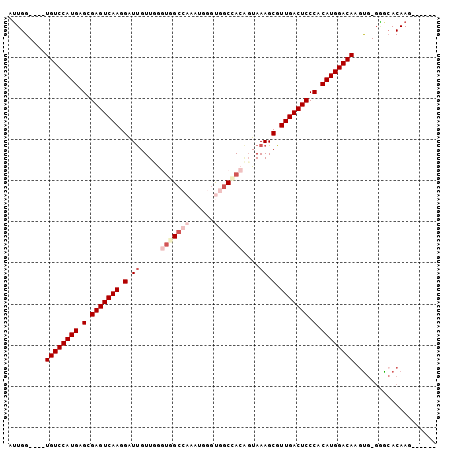
>X_DroMel_CAF1 1546165 93 - 22224390 AUUGG----UGUCCAUGAGCGAGUCAAGGAUUGUUUGGUGGCCAAAUGGGUGGCAACAGUAAAGCGUUGACUCCCACAUGGACAAUUG-UGGCACAAG------ .....----((((((((.(.(((((((.(.....(((...((((......))))..))).....).))))))).).))))))))....-.........------ ( -32.90) >DroSec_CAF1 8546 93 - 1 AUUGG----UGUCCAUGAGCGAGUCAAGGGUUGUUUGGUGGCCAAAUGGGUGGCCACAGUAAAGCGUUGACUCCCACAUGGACAAGUG-GAGCACAAG------ .....----((((((((.(.(((((((.(.((.....(((((((......)))))))....)).).))))))).).)))))))).(((-...)))...------ ( -40.30) >DroSim_CAF1 9565 93 - 1 AUUGG----UGUCCAUGAGCGAGUCAAGGGUUGUUUGGUGGCCAAAUGGGUGGCUACAGUAAAGCGUUGACUCCCACAUGGACAAGUG-GAGCACAAG------ .....----((((((((.(.(((((((.(.((.....(((((((......)))))))....)).).))))))).).)))))))).(((-...)))...------ ( -38.00) >DroEre_CAF1 10103 93 - 1 AUUGG----UGUCCAUGAGCGAGUCAAGGAUUGUUGAUUGGCCAACUGGGUGGCCACAGUAAAGCGUUGACUCCCACAUGGACAAGUG-GGACACAAG------ .....----((((((((.(.(((((((.(....(((..((((((......))))))...)))..).))))))).).)))))))).(((-...)))...------ ( -35.40) >DroYak_CAF1 8749 90 - 1 AUUGG----UGUCCAUGAGCGAGUCAAGGAUUGUUGAUUGGC---CUGGGUGGCCACGGUAAAGCGUUGACUCCCACAUGGACAAGUG-GGACACAAG------ .....----((((((((.(.(((((((.(.(((..(..((((---(.....))))))..)))..).))))))).).)))))))).(((-...)))...------ ( -34.80) >DroPer_CAF1 12688 104 - 1 AGUGGGUCAUGUCCAUGAGCGAGUCAAGGAUCGUUGGGGAGCCUCCACUUCCUCCACUCUAAAGCGUUGACUCCCACAUGGACAUGCACGCGGACAAGCGGAAU .....(.((((((((((.(.(((((((.(...((.((((((.......)))))).)).......).))))))).).))))))))))).(((......))).... ( -38.30) >consensus AUUGG____UGUCCAUGAGCGAGUCAAGGAUUGUUGGGUGGCCAAAUGGGUGGCCACAGUAAAGCGUUGACUCCCACAUGGACAAGUG_GGGCACAAG______ .........((((((((.(.(((((((.(.((.....(((((((......)))))))....)).).))))))).).)))))))).................... (-29.87 = -31.62 + 1.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:16 2006