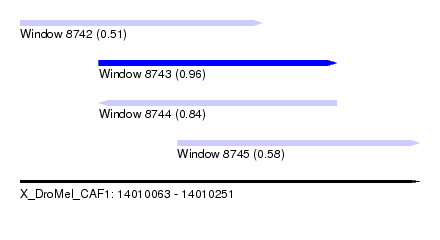
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,010,063 – 14,010,251 |
| Length | 188 |
| Max. P | 0.964322 |

| Location | 14,010,063 – 14,010,177 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.17 |
| Mean single sequence MFE | -26.34 |
| Consensus MFE | -18.18 |
| Energy contribution | -17.58 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14010063 114 + 22224390 ACA---UACAUCAAUGUUUUAACUAAGUAUUAGCGUUAAAAAAUAUAAUAUAAAGCUUUAAUUAAGU---UGGGCUUUUUAUAAACAGCUUGCAGCUUUGAUUAAACUUGGAAGCUGCGA ...---..........(((((((...........)))))))......(((.(((((((.........---.))))))).))).......((((((((((.(.......).)))))))))) ( -21.20) >DroSec_CAF1 36743 114 + 1 ACA---UGAAUCAAUGUUUGAACUAAGUAUUAGCGUUAGCAAAUAUAAUAUAAAGCUUUUAUUAAGU---UGGGCUUUUUAUAAACAGCUUGCAGCUUUGAUUAAACUUGGAAACUGCGA ...---....((((.((((((.....(((((.((....)).)))))....(((((((.....(((((---((.............))))))).))))))).))))))))))......... ( -20.22) >DroSim_CAF1 38180 114 + 1 ACA---CGAAUCAAUGUUUUAACUAAGUAUUAGCGUUAGCAAAUAUAAUAUAAAGCUUUGAUUAAGU---UGGGCUUUUUAUGAACAGCUUGCAGCUUUGAUUAAACUUGGAAGCUGCGA ...---..((((((.((((((.....(((((.((....)).)))))....)))))).))))))((((---((..(.......)..))))))((((((((.(.......).)))))))).. ( -31.60) >DroEre_CAF1 34717 114 + 1 ACA---CGCAUCAAUGU-UAAAAAAACGAUUAGCGCUA--CAAUGUAAUGUGAAGCUUUGAUUAAGCCGCUGGGCUUUUUAUGAACAGCUUGCAGCUUUGAUGAAACUUGGAAGCUGCGA ...---(((((....((-(((........)))))((..--....)).)))))(((((((.((.(((((....)))))...)).)).)))))((((((((.(.......).)))))))).. ( -29.50) >DroYak_CAF1 37474 118 + 1 ACAAUAAGCAUCGAUUUUUUAAAUAACGAUUAGCGCUA--AAAUGUAAUAAAAAGCUUUGAUUAAGUCGCUGGGCUUUUUAUGAGCAGCUUGCAGCUUUGAUUAAACUUGGAAGCUGCGA .....((((((((.((........))))))....(((.--.......(((((((((((((((...))))..))))))))))).))).))))((((((((.(.......).)))))))).. ( -29.20) >consensus ACA___CGCAUCAAUGUUUUAACUAAGUAUUAGCGUUA__AAAUAUAAUAUAAAGCUUUGAUUAAGU___UGGGCUUUUUAUGAACAGCUUGCAGCUUUGAUUAAACUUGGAAGCUGCGA ...............................(((((........)).(((.(((((((.............))))))).))).....)))(((((((((.(.......).))))))))). (-18.18 = -17.58 + -0.60)


| Location | 14,010,100 – 14,010,212 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.41 |
| Mean single sequence MFE | -27.06 |
| Consensus MFE | -22.32 |
| Energy contribution | -21.72 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14010100 112 + 22224390 AAAUAUAAUAUAAAGCUUUAAUUAAGU---UGGGCUUUUUAUAAACAGCUUGCAGCUUUGAUUAAACUUGGAAGCUGCGACUAUUUUAAGCAUUU-UCAAAGAGCUUUUAGUUUAA---- .....(((...((((((((....((((---...((((.....(((.((.((((((((((.(.......).)))))))))))).))).))))))))-....))))))))....))).---- ( -24.20) >DroSec_CAF1 36780 113 + 1 AAAUAUAAUAUAAAGCUUUUAUUAAGU---UGGGCUUUUUAUAAACAGCUUGCAGCUUUGAUUAAACUUGGAAACUGCGACUAUUUUAAGCAUUUUUCCAAGAGCUUUUAGUUUAA---- .....(((...((((((((((..((((---((((((.((....)).))))..))))))....))))((((((((.(((...........)))..))))))))))))))....))).---- ( -23.20) >DroSim_CAF1 38217 112 + 1 AAAUAUAAUAUAAAGCUUUGAUUAAGU---UGGGCUUUUUAUGAACAGCUUGCAGCUUUGAUUAAACUUGGAAGCUGCGACUAUUUUAAGCAUUU-UGCAAGAGCUUUUAGUUUAA---- .....(((...((((((((....((((---((..(.......)..))))))((((((((.(.......).))))))))...........((....-.)).))))))))....))).---- ( -27.60) >DroEre_CAF1 34751 115 + 1 CAAUGUAAUGUGAAGCUUUGAUUAAGCCGCUGGGCUUUUUAUGAACAGCUUGCAGCUUUGAUGAAACUUGGAAGCUGCGACUAUUUCAAGCAUUU-UCCAAAAGCUUUUAGUGUAA---- ...........((((((((((..(.((.((((..(.......)..))))((((((((((.(.......).)))))))))).........)).)..-))..))))))))........---- ( -29.20) >DroYak_CAF1 37512 119 + 1 AAAUGUAAUAAAAAGCUUUGAUUAAGUCGCUGGGCUUUUUAUGAGCAGCUUGCAGCUUUGAUUAAACUUGGAAGCUGCGACUAUUUUAAGCAUAU-UCCAAAAGCUUUUGGUUUAGUUUU ..............((((.((...(((((((..((((.....)))))))..((((((((.(.......).))))))))))))..)).)))).((.-.(((((....)))))..))..... ( -31.10) >consensus AAAUAUAAUAUAAAGCUUUGAUUAAGU___UGGGCUUUUUAUGAACAGCUUGCAGCUUUGAUUAAACUUGGAAGCUGCGACUAUUUUAAGCAUUU_UCCAAGAGCUUUUAGUUUAA____ ...........((((((((..............((((.....(((.((.((((((((((.(.......).)))))))))))).))).)))).........))))))))............ (-22.32 = -21.72 + -0.60)



| Location | 14,010,100 – 14,010,212 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.41 |
| Mean single sequence MFE | -21.39 |
| Consensus MFE | -15.98 |
| Energy contribution | -15.86 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.839319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14010100 112 - 22224390 ----UUAAACUAAAAGCUCUUUGA-AAAUGCUUAAAAUAGUCGCAGCUUCCAAGUUUAAUCAAAGCUGCAAGCUGUUUAUAAAAAGCCCA---ACUUAAUUAAAGCUUUAUAUUAUAUUU ----........((((((..((((-....((((.(((((((.(((((((.............)))))))..))))))).....))))...---..))))....))))))........... ( -19.92) >DroSec_CAF1 36780 113 - 1 ----UUAAACUAAAAGCUCUUGGAAAAAUGCUUAAAAUAGUCGCAGUUUCCAAGUUUAAUCAAAGCUGCAAGCUGUUUAUAAAAAGCCCA---ACUUAAUAAAAGCUUUAUAUUAUAUUU ----........((((((.(((.......((((.(((((((.(((((((.............)))))))..))))))).....))))...---......))).))))))........... ( -18.21) >DroSim_CAF1 38217 112 - 1 ----UUAAACUAAAAGCUCUUGCA-AAAUGCUUAAAAUAGUCGCAGCUUCCAAGUUUAAUCAAAGCUGCAAGCUGUUCAUAAAAAGCCCA---ACUUAAUCAAAGCUUUAUAUUAUAUUU ----........((((((...((.-....))....((((((.(((((((.............)))))))..)))))).............---..........))))))........... ( -18.12) >DroEre_CAF1 34751 115 - 1 ----UUACACUAAAAGCUUUUGGA-AAAUGCUUGAAAUAGUCGCAGCUUCCAAGUUUCAUCAAAGCUGCAAGCUGUUCAUAAAAAGCCCAGCGGCUUAAUCAAAGCUUCACAUUACAUUG ----.........(((((((((((-(..(((...........)))..)))))..........(((((((..(((..........)))...)))))))....)))))))............ ( -25.50) >DroYak_CAF1 37512 119 - 1 AAAACUAAACCAAAAGCUUUUGGA-AUAUGCUUAAAAUAGUCGCAGCUUCCAAGUUUAAUCAAAGCUGCAAGCUGCUCAUAAAAAGCCCAGCGACUUAAUCAAAGCUUUUUAUUACAUUU .........(((((....)))))(-(((.((((.....(((((((((((...(((((.....)))))..)))))(((.......)))...))))))......))))....))))...... ( -25.20) >consensus ____UUAAACUAAAAGCUCUUGGA_AAAUGCUUAAAAUAGUCGCAGCUUCCAAGUUUAAUCAAAGCUGCAAGCUGUUCAUAAAAAGCCCA___ACUUAAUCAAAGCUUUAUAUUAUAUUU ............((((((...........(((......))).(((((((...(((((.....)))))..)))))))...........................))))))........... (-15.98 = -15.86 + -0.12)



| Location | 14,010,137 – 14,010,251 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.40 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -18.56 |
| Energy contribution | -19.16 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14010137 114 + 22224390 AUAAACAGCUUGCAGCUUUGAUUAAACUUGGAAGCUGCGACUAUUUUAAGCAUUU-UCAAAGAGCUUUUAGUUUAA-----AUAUAGCAUGAAACUUUAUGGUUUUCCUAGUUUUAACAC .......(((.((((((((.(.......).))))))))(((((....((((....-.......)))).)))))...-----....))).(((((((....((....)).))))))).... ( -25.60) >DroSec_CAF1 36817 115 + 1 AUAAACAGCUUGCAGCUUUGAUUAAACUUGGAAACUGCGACUAUUUUAAGCAUUUUUCCAAGAGCUUUUAGUUUAA-----AUAUAGCAUGCAACUUUAGGGUUUUCCUAUUUUAAUAAC ......((.((((((((..(((((((((((((((.(((...........)))..))))))))....)))))))...-----....))).))))))).((((.....)))).......... ( -22.70) >DroSim_CAF1 38254 114 + 1 AUGAACAGCUUGCAGCUUUGAUUAAACUUGGAAGCUGCGACUAUUUUAAGCAUUU-UGCAAGAGCUUUUAGUUUAA-----AUAUAGCAUGAAACUUUAGGGUUUUCCUAUUUUAAUUAC .((((.(((((((((((((.(.......).))))))))...........((....-.))..)))))))))..((((-----(.((((...((((((....)))))).))))))))).... ( -24.70) >DroEre_CAF1 34791 114 + 1 AUGAACAGCUUGCAGCUUUGAUGAAACUUGGAAGCUGCGACUAUUUCAAGCAUUU-UCCAAAAGCUUUUAGUGUAA-----AUGUAGCAUAAAACUUGAUAGUUUCCUUAGCUUCAACAC .(((..((((.((((((((.(.......).))))))))(((((((..((((....-.......))))...((((..-----.....)))).......))))))).....))))))).... ( -23.40) >DroYak_CAF1 37552 119 + 1 AUGAGCAGCUUGCAGCUUUGAUUAAACUUGGAAGCUGCGACUAUUUUAAGCAUAU-UCCAAAAGCUUUUGGUUUAGUUUUAGCUUAGCAUAAAACUUGAAUGUUUUCUUAGUUUAGACUC .(((((((.((((((((((.(.......).)))))))))))).....((((.((.-.(((((....)))))..))))))..))))).....(((((.(((....)))..)))))...... ( -29.00) >consensus AUGAACAGCUUGCAGCUUUGAUUAAACUUGGAAGCUGCGACUAUUUUAAGCAUUU_UCCAAGAGCUUUUAGUUUAA_____AUAUAGCAUGAAACUUUAGGGUUUUCCUAGUUUAAACAC .......(((.((((((((.(.......).))))))))(((((....((((............)))).)))))............)))..((((((....)))))).............. (-18.56 = -19.16 + 0.60)

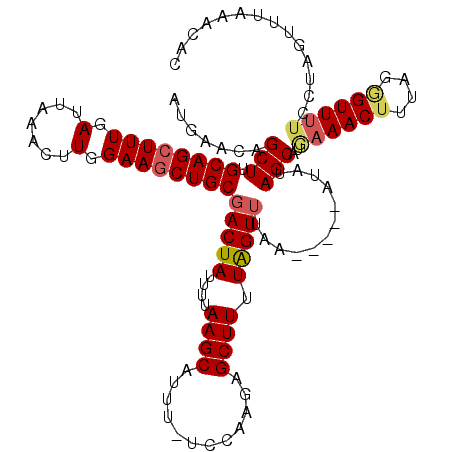

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:47 2006