| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,003,640 – 14,003,752 |
| Length | 112 |
| Max. P | 0.523653 |

| Location | 14,003,640 – 14,003,752 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 83.34 |
| Mean single sequence MFE | -39.73 |
| Consensus MFE | -27.33 |
| Energy contribution | -28.20 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
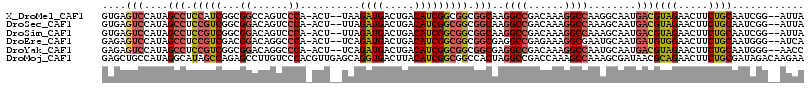
>X_DroMel_CAF1 14003640 112 - 22224390 GUGAGUCCAUAGCCUCCAUCGGCGGCCAGUCCCA-ACU--UAAGAUGACUGACAUCGGCGGCGGCAAGGCCGACAAAGGCCAAGGCAAUGACGUAGAACUUCUGCAAUCGG--AUUA ...(((((...((((....(.((.((((((....-)))--...((((.....))))))).)).)...((((......)))).))))......((((.....))))....))--))). ( -37.10) >DroSec_CAF1 30891 112 - 1 GUGAGUCCAUAGCCUCCGUCGGCGGACAGUCCCA-ACU--UUAGAUGACUGACAUCGGCGGCGGCAAGGCCGACAAAGGCCAAAGCAAUGACGUAGAACUUCUGCAAUCGG--AUUA ...(((((...(((.((((((..(..(((((...-...--......))))).)..)))))).)))..((((......))))...........((((.....))))....))--))). ( -41.30) >DroSim_CAF1 31383 112 - 1 GUGAGUCCAUAGCCUCCGUCGGCGGACAGUCCCA-ACU--UUAGAUGACUGACAUCGGCGGCGGCAAGGCCGACAAAGGCCAAAGCAAUGACGUAGAACUUCUGCAAUCGG--AUUA ...(((((...(((.((((((..(..(((((...-...--......))))).)..)))))).)))..((((......))))...........((((.....))))....))--))). ( -41.30) >DroEre_CAF1 30402 112 - 1 GAGAGUCCAUAGCCUCCGUCGACGGACAGGCCCA-ACU--UCAGAUGACUGACAUCGGCGGCGGCGAGGCCGAGAAAGGCGAAUGCAAUGAUGUGGAACUUCUGCAAUGGG--AUCA ..((..((((.(((.(((((((.((......)).-...--((((....))))..))))))).)))...(((......)))...........((..(.....)..)))))).--.)). ( -37.70) >DroYak_CAF1 32301 112 - 1 GAGAGUCCAUAGCCUCCGUCGGCGGACAGGCCCA-ACU--UCAGAUGACUGACAUCGGCGGCGGCGAGGCCGACAAAGGCCAAUGCAAUGACGUAGAACUUCUGCAAUGGG--AACC .....(((((.(((.((((((((......)))..-...--...((((.....))))))))).)))..((((......))))...........((((.....)))).)))))--.... ( -39.50) >DroMoj_CAF1 47355 117 - 1 GAGCUGCCAUAGGCAUAGCCAGAGCCUUGUCCCACGUUGAGCAGGUGACUUACAUCGGCGGCCACUAGGCCGACCAAAGCCAAAGCGAUAACGCAGAACUUCUGCGAUAGACAAGAA (.((((((...)))..)))).....((((((...((((..((.((((.....))))(((((((....))))).))...))...))))....(((((.....)))))...)))))).. ( -41.50) >consensus GAGAGUCCAUAGCCUCCGUCGGCGGACAGUCCCA_ACU__UCAGAUGACUGACAUCGGCGGCGGCAAGGCCGACAAAGGCCAAAGCAAUGACGUAGAACUUCUGCAAUCGG__AUUA ....(((....(((.(((((...((......))..........((((.....))))))))).)))..((((......))))........)))((((.....))))............ (-27.33 = -28.20 + 0.87)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:42 2006