| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,003,011 – 14,003,105 |
| Length | 94 |
| Max. P | 0.745186 |

| Location | 14,003,011 – 14,003,105 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 76.04 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -16.15 |
| Energy contribution | -17.07 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
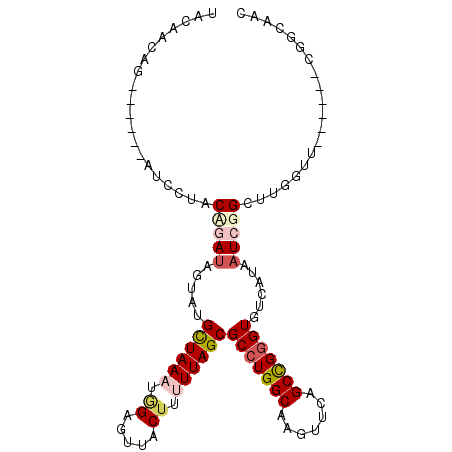
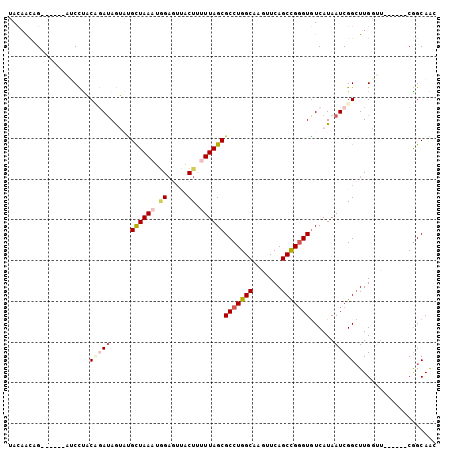
>X_DroMel_CAF1 14003011 94 - 22224390 UACUACAGUUGUACAUCCUACAGAUAGUAUGCUAAAGCG-----CUUUUUAGCGCCUGGCAAGUUCAGCUGGGUGUCAUAAUCGGCUUGGUC------CGGCAAC (((((...(((((.....))))).)))))(((((..(((-----((....))))).)))))......((((((.(((......)))....))------))))... ( -27.80) >DroPse_CAF1 135661 97 - 1 GAUAGUAG--------CUCUCUAAUAAGUCGCUAAAUGGAGUCACUGAUUAGCGCCUGGCAAGUUCAGCCGGGUGUCAUAAUCGGCUUGCAUUGGCAACGGCAGC ..(((..(--------((........)))..)))......(((.((((((((((((((((.......)))))))))..))))))).((((....)))).)))... ( -32.10) >DroSim_CAF1 30729 94 - 1 UACAACUGU-----AUCCUACCGAUAGUAUGCUAAAUGGAGUUACUUUUUAGCGCCUGGCAAGUUCAGCUGGGUAUCAUAAUCGGCUUGGUU------CGGCAAC .....(((.-----..((..(((((.....((((((.((.....)).))))))(((..((.......))..)))......)))))...))..------))).... ( -26.00) >DroEre_CAF1 29791 94 - 1 UACAGCUGA-----AUCCUACAGAUAGUAUGUUAAAUGGAGUUACUUUUUAGCGCCUGGCAAGUUCAGCCGGGUGUCAUAAUCGGAUUGAUU------CAGCAAC ....(((((-----(((.....(((.....((((((.((.....)).))))))(((((((.......)))))))......))).....))))------))))... ( -30.20) >DroYak_CAF1 31669 94 - 1 UACAACUGA-----AUCCUACCGAGAGUAUGCUAAAUAGAGUUACUUUUUAGCGCCUGGCAAGUUCAGCCGUGUGUCAUAAUCGGCUUGGUU------CGGCAAC .....((((-----(((...((((...(((((((((.((.....)).))))))((.((((.......)))).))...))).))))...))))------))).... ( -25.10) >DroPer_CAF1 156435 97 - 1 GAUAGUAG--------CUCUCUAAUAAGUCGCUAAAUGGAGUCACUGAUUAGCGCCUGGCAAGUUCAGCCGGGUGUCAUAAUCGGCUUGCAUUGGCGACGGCAGC .......(--------((.........((((((((.(((((((...((((((((((((((.......)))))))))..)))))))))).)))))))))))))... ( -35.20) >consensus UACAACAG______AUCCUACAGAUAGUAUGCUAAAUGGAGUUACUUUUUAGCGCCUGGCAAGUUCAGCCGGGUGUCAUAAUCGGCUUGGUU______CGGCAAC ....................(((((.....((((((.((.....)).))))))(((((((.......)))))))......))))).................... (-16.15 = -17.07 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:41 2006