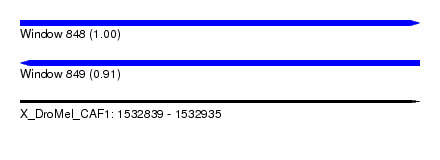
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,532,839 – 1,532,935 |
| Length | 96 |
| Max. P | 0.996261 |

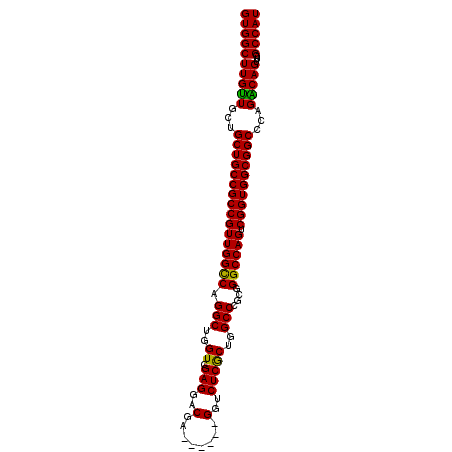
| Location | 1,532,839 – 1,532,935 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 94.28 |
| Mean single sequence MFE | -58.38 |
| Consensus MFE | -56.24 |
| Energy contribution | -55.42 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1532839 96 + 22224390 GUGGCUUGCUGCUGCUGCCGCCGUUGGCCAGGCUGGUCGAGGACGA------GGUCUCGCUGGCCCGCGGGCCAGUCGGUGGCGGCCCAGGCAGUUCGCCAU (((((..((((((((((((((((((((((.(((..(.(((((....------..))))))..)))....)))))).))))))))))...))))))..))))) ( -63.20) >DroSec_CAF1 5822 96 + 1 GUGGCUUGUUGCUGCUGCCGCCGUUGGUCAGGCUGGUCGAGGACGA------GGUCUCGCUGGCCCGCGGGCCAGUCGGUGGCGGCCCAGGCAGUUCGCCAU (((((..((((((((((((((((((((((.(((..(.(((((....------..))))))..)))....)))))).))))))))))...))))))..))))) ( -55.20) >DroEre_CAF1 5722 96 + 1 GUGGCUUGUUGCUGCUGCCGCCGUUGGCCAGGCUGGUCGAGGACGA------GGUCUCGCUGGCCCGCGGGCCAGUCGGUGGCGGCCCAGACAGUUCGCCAU ((((((((((...((((((((((((((((.(((..(.(((((....------..))))))..)))....)))))).))))))))))...)))))...))))) ( -56.80) >DroYak_CAF1 5695 102 + 1 GUGGCUUGUUGCUGCUGCCGCCGUUGGCCAGGCUGGUCGAGGACGAGGUUGAGGUCUCACUGGCCCGCGGGCCAGUCGGUGGCGGCACAGACAGUUCGCCAU ((((((((((..(((((((((((((((((.(((..((.(((..(........)..)))))..)))....)))))).)))))))))))..)))))...))))) ( -58.30) >consensus GUGGCUUGUUGCUGCUGCCGCCGUUGGCCAGGCUGGUCGAGGACGA______GGUCUCGCUGGCCCGCGGGCCAGUCGGUGGCGGCCCAGACAGUUCGCCAU ((((((((((...((((((((((((((((.(((..((.(((..(........)..)))))..)))....)))))).))))))))))...)))))...))))) (-56.24 = -55.42 + -0.81)

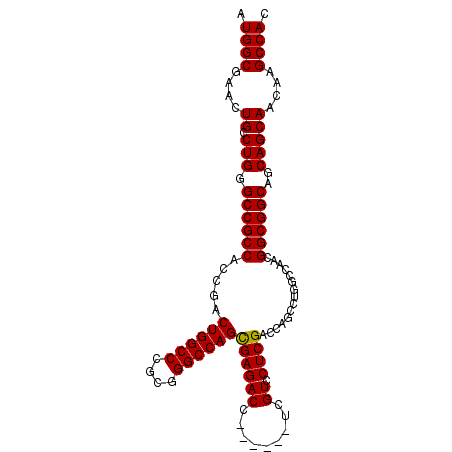
| Location | 1,532,839 – 1,532,935 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 94.28 |
| Mean single sequence MFE | -43.27 |
| Consensus MFE | -40.39 |
| Energy contribution | -40.20 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1532839 96 - 22224390 AUGGCGAACUGCCUGGGCCGCCACCGACUGGCCCGCGGGCCAGCGAGACC------UCGUCCUCGACCAGCCUGGCCAACGGCGGCAGCAGCAGCAAGCCAC .((((...((((.((.((((((.....((((((....))))))((((...------.....))))...............))))))..))))))...)))). ( -45.10) >DroSec_CAF1 5822 96 - 1 AUGGCGAACUGCCUGGGCCGCCACCGACUGGCCCGCGGGCCAGCGAGACC------UCGUCCUCGACCAGCCUGACCAACGGCGGCAGCAGCAACAAGCCAC .((((....(((.((.((((((.....((((((....))))))((((...------.....))))...............))))))..)))))....)))). ( -42.00) >DroEre_CAF1 5722 96 - 1 AUGGCGAACUGUCUGGGCCGCCACCGACUGGCCCGCGGGCCAGCGAGACC------UCGUCCUCGACCAGCCUGGCCAACGGCGGCAGCAGCAACAAGCCAC .((((....((.(((.((((((.....((((((....))))))((((...------.....))))...............))))))..)))))....)))). ( -42.00) >DroYak_CAF1 5695 102 - 1 AUGGCGAACUGUCUGUGCCGCCACCGACUGGCCCGCGGGCCAGUGAGACCUCAACCUCGUCCUCGACCAGCCUGGCCAACGGCGGCAGCAGCAACAAGCCAC .((((....((.((((((((((..(((((((((....)))))))(((........))).....)).((.....)).....))))))).)))))....)))). ( -44.00) >consensus AUGGCGAACUGCCUGGGCCGCCACCGACUGGCCCGCGGGCCAGCGAGACC______UCGUCCUCGACCAGCCUGGCCAACGGCGGCAGCAGCAACAAGCCAC .((((....((.(((.((((((.....((((((....))))))((((((.........)).))))...............))))))..)))))....)))). (-40.39 = -40.20 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:14 2006