
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,984,145 – 13,984,297 |
| Length | 152 |
| Max. P | 0.565671 |

| Location | 13,984,145 – 13,984,264 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.18 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -15.70 |
| Energy contribution | -18.54 |
| Covariance contribution | 2.84 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13984145 119 - 22224390 GGGAUACUGGGUACUGGAUUGUGGACUCUGUCGGUGGCUACCUGCUUGUGCCUCAGCUCAGAAUUCGGCAGCAAUUUUUAUCCAGCGUUCCUCUUGCACCGUUUUCCACCAUAUAUAUA ((((..(((((((..((((((((((.((((...(.(((.((......))))).)....)))).)))....))))))).)))))))..))))............................ ( -30.70) >DroSec_CAF1 17339 109 - 1 -------UGGAUACUGGAUUGUGGAUUCUGUCUGUGGCUACCUGCUUGUGCCGCAGCUCAGAAUUCGGCAGCAGUUUUUAUCCAGCGUUCCUCUUGCACCGUUUUCCACCAUAUAU--- -------((((((..(((((((((((((((.(((((((.((......)))))))))..))))))))....))))))).))))))(((.......)))...................--- ( -34.70) >DroSim_CAF1 17423 96 - 1 -------UGGAUACUGGAUUG-------------UGGCUACCUGCUUGUGCCGCAGCUCAGAAUUCGGCAGCAAUUUUUAUCCAGCGUUGCUCUUGCACCGUUUUCCACCAUAUAU--- -------((((..((((((((-------------((((.((......))))))))((((.(....).).))).......))))))((.(((....))).))...))))........--- ( -25.80) >DroEre_CAF1 11856 104 - 1 G-GAU-------ACUGGAUUGUGGGUUCUGGCUGUGGCUACCA-C--UUGCCGCAGCUCAGAAUUCGGCAGCACAUUUUAUCCAGCGCUCCUCUUCCACCGUU-UCCACCAUAUAU--- (-((.-------((((((....((((((((((((((((.....-.--..)))))))).))))))))((.(((.(..........).)))))...))))..)).-))).........--- ( -39.30) >DroYak_CAF1 18066 112 - 1 G-GAUACUGCAUACUGGAUUCUGGAUACUGUUUGUGGCUGCCA-C--UUGCCGCAACUCAGAAUUCGGCAGCAAUUUUUAUCCAGCGCUUCGCUUCCUCCGUUUUCCAGCAUAUAU--- (-((((.(((...(((((((((((.....(((.(((((.....-.--..)))))))))))))))))))..))).....)))))((((...))))......................--- ( -30.00) >consensus G_GAU__UGGAUACUGGAUUGUGGAUUCUGUCUGUGGCUACCUGCUUGUGCCGCAGCUCAGAAUUCGGCAGCAAUUUUUAUCCAGCGUUCCUCUUGCACCGUUUUCCACCAUAUAU___ .............((((((..(((((((((.(((((((...........)))))))..)))))))))............)))))).................................. (-15.70 = -18.54 + 2.84)


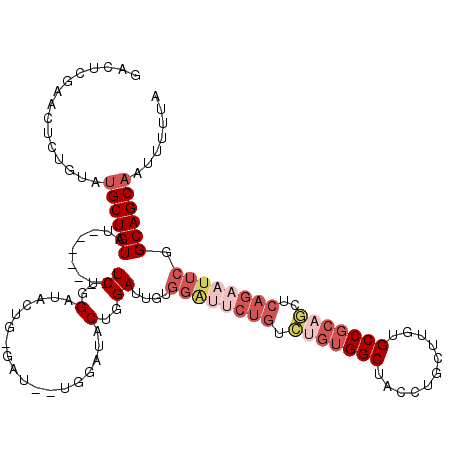
| Location | 13,984,184 – 13,984,297 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.22 |
| Mean single sequence MFE | -34.52 |
| Consensus MFE | -18.53 |
| Energy contribution | -21.37 |
| Covariance contribution | 2.84 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13984184 113 - 22224390 GACUCGAACUCUGUAUGCUGUAU-------UCUGGAUACUGGGAUACUGGGUACUGGAUUGUGGACUCUGUCGGUGGCUACCUGCUUGUGCCUCAGCUCAGAAUUCGGCAGCAAUUUUUA ...(((((.((((...(((((((-------((..(...)..)))))).((((((.((...((((.((........))))))...)).)))))).))).)))).)))))............ ( -31.80) >DroSec_CAF1 17375 105 - 1 GACUCGAACUCUGUGUGCUGUAU-------UCUGGAUAC--------UGGAUACUGGAUUGUGGAUUCUGUCUGUGGCUACCUGCUUGUGCCGCAGCUCAGAAUUCGGCAGCAGUUUUUA .....(((..((((.((((((((-------((.......--------.))))))........((((((((.(((((((.((......)))))))))..))))))))))))))))..))). ( -36.40) >DroSim_CAF1 17459 92 - 1 GACUCGAACUCUGUGUGCUGUCU-------UCUGGAUAC--------UGGAUACUGGAUUG-------------UGGCUACCUGCUUGUGCCGCAGCUCAGAAUUCGGCAGCAAUUUUUA ...............(((((((.-------((..(...)--------..))..(((..(((-------------((((.((......)))))))))..))).....)))))))....... ( -28.20) >DroEre_CAF1 11891 109 - 1 GACUCGAACUCUGUACGCUGUACGCUGUAUUCUGGAUACUG-GAU-------ACUGGAUUGUGGGUUCUGGCUGUGGCUACCA-C--UUGCCGCAGCUCAGAAUUCGGCAGCACAUUUUA ................(((((.....((((((........)-)))-------)).......(((((((((((((((((.....-.--..)))))))).))))))))))))))........ ( -40.00) >DroYak_CAF1 18102 116 - 1 GACUCGAACUCUGUAUGCUGUAUGCUGUACUCUUGAUACUG-GAUACUGCAUACUGGAUUCUGGAUACUGUUUGUGGCUGCCA-C--UUGCCGCAACUCAGAAUUCGGCAGCAAUUUUUA ...............((((((((((.(((..(........)-..))).)))))(((((((((((.....(((.(((((.....-.--..))))))))))))))))))))))))....... ( -36.20) >consensus GACUCGAACUCUGUAUGCUGUAU_______UCUGGAUACUG_GAU__UGGAUACUGGAUUGUGGAUUCUGUCUGUGGCUACCUGCUUGUGCCGCAGCUCAGAAUUCGGCAGCAAUUUUUA ...............((((((.........((..(..................)..))....((((((((.(((((((...........)))))))..)))))))).))))))....... (-18.53 = -21.37 + 2.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:35 2006