
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,980,269 – 13,980,498 |
| Length | 229 |
| Max. P | 0.938624 |

| Location | 13,980,269 – 13,980,379 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 89.01 |
| Mean single sequence MFE | -24.88 |
| Consensus MFE | -21.34 |
| Energy contribution | -21.70 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13980269 110 - 22224390 GCAUCGGGCGCGCAAAUAAAUUAGCGGCCACCAAGUGUUGACAUUUAUUGUCAAGCCAAGCCCCAAAAAAAGGCAAACUGAGAGAACGCGAGUUCGAAACCGA-----AACCAAA ...((((...(.........((((.((((((...)))((((((.....)))))))))..(((.........)))...))))..((((....)))))...))))-----....... ( -27.60) >DroPse_CAF1 118072 92 - 1 GCAUCGAGCGCGCAAAUAAAUUAGCGGCCGCAAAGUGUUGACAUUUAUUGUCAAGCCAAGCCCCAAAAAAA-G-AAACAGA---------------------AACCGAAACGGAA ((..((.((.(((..........)))))))....(..((((((.....))))))..)..))..........-.-.......---------------------..(((...))).. ( -15.50) >DroSec_CAF1 13334 109 - 1 GCAUCGGGCGCGCAAAUAAAUUAGCGGCCACCAAGUGUUGACAUUUAUUGUCAAGCCAAGCCCCAAAAAAA-GCAAACUGAGGGAACGCGAGUUCGAAACCGA-----AACCAAA ...((((((.(((............((((((...)))((((((.....)))))))))...(((((......-......)).)))...))).))))))......-----....... ( -26.80) >DroSim_CAF1 13413 109 - 1 GCAUCGGGCGCGCAAAUAAAUUAGCGGCCACCAAGUGUUGACAUUUAUUGUCAAGCCAAGCCCCAAAAAAA-GCAAACUGAGAGAACGCGAGUUCGAAACCGA-----AACCAAA ...((((((((............))((((((...)))((((((.....)))))))))..))))........-...........((((....))))))......-----....... ( -26.50) >DroEre_CAF1 7730 108 - 1 GCAUCG-GCGCGCAAAUAAAUUAGCGGCCACCAAGUGUUGACAUUUAUUGUCAAGCCAAGCCCCAAAAAAA-GCAAACUGAGAGAACGCGAGUUCGAAACCGA-----AACCAAA ...(((-(.((((..........))((((((...)))((((((.....)))))))))..............-)).........((((....))))....))))-----....... ( -24.90) >DroYak_CAF1 14092 109 - 1 GCAUCGGGCGCGCAAAUAAAUUAGCGGCCACCAAGUGUUGACAUUUAUUGUCAAGCCAAGCCCCAAAAAAA-GCAAACUGCGAGAACGCGAGUUCGAAACCGA-----AACCAAA ...((((((((............))((((((...)))((((((.....)))))))))..))))........-((.....))))((((....))))........-----....... ( -28.00) >consensus GCAUCGGGCGCGCAAAUAAAUUAGCGGCCACCAAGUGUUGACAUUUAUUGUCAAGCCAAGCCCCAAAAAAA_GCAAACUGAGAGAACGCGAGUUCGAAACCGA_____AACCAAA .....((((((............))((((((...)))((((((.....)))))))))..))))....................((((....)))).................... (-21.34 = -21.70 + 0.36)

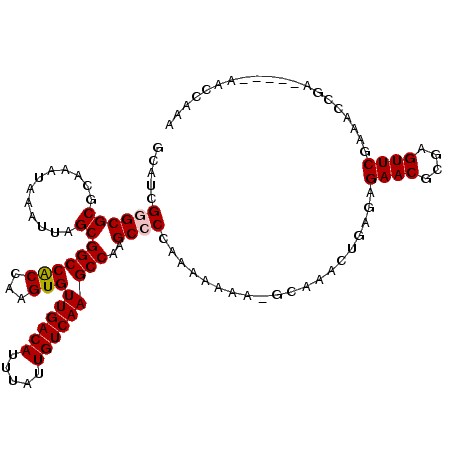
| Location | 13,980,299 – 13,980,418 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 97.80 |
| Mean single sequence MFE | -32.86 |
| Consensus MFE | -29.16 |
| Energy contribution | -29.52 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13980299 119 - 22224390 CCACCUCCUCCUGGGAAACCGCCACCUCAUGUGGCGAACGCAUCGGGCGCGCAAAUAAAUUAGCGGCCACCAAGUGUUGACAUUUAUUGUCAAGCCAAGCCCCAAAAAAAGGCAAACUG ...(((.....((((....((((((.....))))))...((....(((.(((..........)))))).....(..((((((.....))))))..)..)))))).....)))....... ( -34.70) >DroSec_CAF1 13364 118 - 1 CCAGCUCCUCCUGGGAAACCGCCACCUCAUGUGGCGAACGCAUCGGGCGCGCAAAUAAAUUAGCGGCCACCAAGUGUUGACAUUUAUUGUCAAGCCAAGCCCCAAAAAAA-GCAAACUG ...(((.....((((....((((((.....))))))...((....(((.(((..........)))))).....(..((((((.....))))))..)..)))))).....)-))...... ( -35.80) >DroSim_CAF1 13443 118 - 1 CCAGCUCCUCCUGGGAAACCGCCACCUCAUGUGGCGAACGCAUCGGGCGCGCAAAUAAAUUAGCGGCCACCAAGUGUUGACAUUUAUUGUCAAGCCAAGCCCCAAAAAAA-GCAAACUG ...(((.....((((....((((((.....))))))...((....(((.(((..........)))))).....(..((((((.....))))))..)..)))))).....)-))...... ( -35.80) >DroEre_CAF1 7760 117 - 1 CCACCUCCUCCUGGGAAACAGCCACCUCAUGUGGUGAACGCAUCG-GCGCGCAAAUAAAUUAGCGGCCACCAAGUGUUGACAUUUAUUGUCAAGCCAAGCCCCAAAAAAA-GCAAACUG ...........((((.....(((((.....)))))....((...(-((.(((..........)))))).....(..((((((.....))))))..)..))))))......-........ ( -28.80) >DroYak_CAF1 14122 118 - 1 CCACCUCCUCCUGGGAAACAGCCACCUCAUGUGGUGAACGCAUCGGGCGCGCAAAUAAAUUAGCGGCCACCAAGUGUUGACAUUUAUUGUCAAGCCAAGCCCCAAAAAAA-GCAAACUG .....(((.....)))....((((((......))))........((((((............))((((((...)))((((((.....)))))))))..))))........-))...... ( -29.20) >consensus CCACCUCCUCCUGGGAAACCGCCACCUCAUGUGGCGAACGCAUCGGGCGCGCAAAUAAAUUAGCGGCCACCAAGUGUUGACAUUUAUUGUCAAGCCAAGCCCCAAAAAAA_GCAAACUG ...........((((....((((((.....))))))...((....(((.(((..........)))))).....(..((((((.....))))))..)..))))))............... (-29.16 = -29.52 + 0.36)


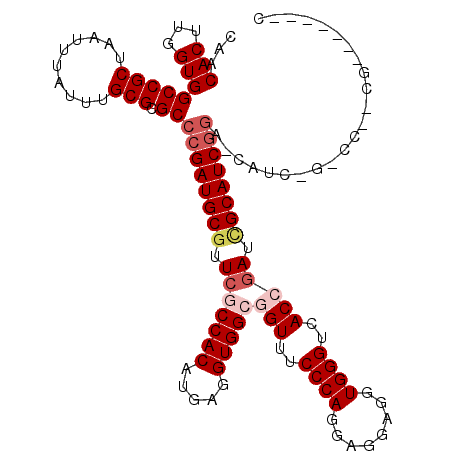
| Location | 13,980,339 – 13,980,458 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.51 |
| Mean single sequence MFE | -44.72 |
| Consensus MFE | -31.28 |
| Energy contribution | -32.72 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.754818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13980339 119 + 22224390 CAACACUUGGUGGCCGCUAAUUUAUUUGCGCGCCCGAUGCGUUCGCCACAUGAGGUGGCGGUUUCCCAGGAGGAGGUGGGUCACCGAUUGCAUCGGACCAUCGGUCCAUCGAAUCGCUC ......((((((((((((((.....))).))..((((((((.(((((((.....)))))(((..((((........))))..))))).))))))))......)).)))))))....... ( -46.90) >DroSec_CAF1 13403 98 + 1 CAACACUUGGUGGCCGCUAAUUUAUUUGCGCGCCCGAUGCGUUCGCCACAUGAGGUGGCGGUUUCCCAGGAGGAGCUGGGCCACCGAUCGCAUCGGG---------------------C ...(((...)))((.((..........))))((((((((((.(((((((.....)))))(((..(((((......)))))..))))).)))))))))---------------------) ( -47.10) >DroSim_CAF1 13482 98 + 1 CAACACUUGGUGGCCGCUAAUUUAUUUGCGCGCCCGAUGCGUUCGCCACAUGAGGUGGCGGUUUCCCAGGAGGAGCUGGGCCACCGAUCGCAUCGGG---------------------C ...(((...)))((.((..........))))((((((((((.(((((((.....)))))(((..(((((......)))))..))))).)))))))))---------------------) ( -47.10) >DroEre_CAF1 7799 117 + 1 CAACACUUGGUGGCCGCUAAUUUAUUUGCGCGC-CGAUGCGUUCACCACAUGAGGUGGCUGUUUCCCAGGAGGAGGUGGGUCACCGAUGGCAUCAGACCAUCGGUCCUCCGAAUCAA-C .....(((((.((((((..........))).))-)((.(((..((((......))))..))).))))))).(((((......(((((((((....).))))))))))))).......-. ( -44.90) >DroYak_CAF1 14161 111 + 1 CAACACUUGGUGGCCGCUAAUUUAUUUGCGCGCCCGAUGCGUUCACCACAUGAGGUGGCUGUUUCCCAGGAGGAGGUGGGUCACCAAUUGCAUCGGAUCAUCAGACCAUCG-------- ......(((((((((((..........))).))((((((((.(((.....)))((((((..(((((.....)))))..).)))))...))))))))..)))))).......-------- ( -37.60) >consensus CAACACUUGGUGGCCGCUAAUUUAUUUGCGCGCCCGAUGCGUUCGCCACAUGAGGUGGCGGUUUCCCAGGAGGAGGUGGGUCACCGAUCGCAUCGGA_CAUC_G_CC__CG_______C ...(((...)))(((((..........))).))((((((((.(((((((.....)))))(((..((((........))))..))))).))))))))....................... (-31.28 = -32.72 + 1.44)

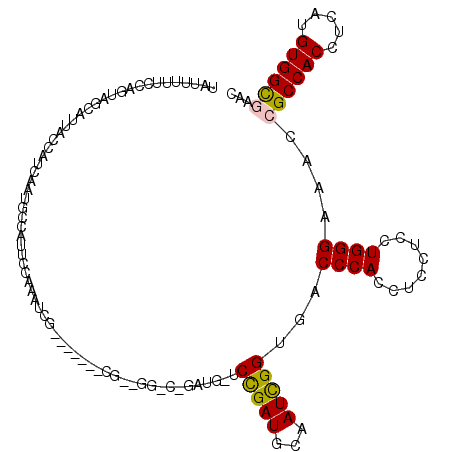
| Location | 13,980,339 – 13,980,458 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.51 |
| Mean single sequence MFE | -43.58 |
| Consensus MFE | -32.60 |
| Energy contribution | -32.80 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13980339 119 - 22224390 GAGCGAUUCGAUGGACCGAUGGUCCGAUGCAAUCGGUGACCCACCUCCUCCUGGGAAACCGCCACCUCAUGUGGCGAACGCAUCGGGCGCGCAAAUAAAUUAGCGGCCACCAAGUGUUG .(((.(((.(.(((.((....(((((((((..(((((..((((........))))..)))(((((.....)))))))..)))))))))((............))))))).).)))))). ( -44.10) >DroSec_CAF1 13403 98 - 1 G---------------------CCCGAUGCGAUCGGUGGCCCAGCUCCUCCUGGGAAACCGCCACCUCAUGUGGCGAACGCAUCGGGCGCGCAAAUAAAUUAGCGGCCACCAAGUGUUG (---------------------(((((((((.(((((..(((((......)))))..)))(((((.....))))))).)))))))))).(((..........))).............. ( -48.60) >DroSim_CAF1 13482 98 - 1 G---------------------CCCGAUGCGAUCGGUGGCCCAGCUCCUCCUGGGAAACCGCCACCUCAUGUGGCGAACGCAUCGGGCGCGCAAAUAAAUUAGCGGCCACCAAGUGUUG (---------------------(((((((((.(((((..(((((......)))))..)))(((((.....))))))).)))))))))).(((..........))).............. ( -48.60) >DroEre_CAF1 7799 117 - 1 G-UUGAUUCGGAGGACCGAUGGUCUGAUGCCAUCGGUGACCCACCUCCUCCUGGGAAACAGCCACCUCAUGUGGUGAACGCAUCG-GCGCGCAAAUAAAUUAGCGGCCACCAAGUGUUG (-(((.(((((((((..((((((.....))))))((((...))))))))))...))).))))((((......))))(((((...(-((.(((..........)))))).....))))). ( -42.10) >DroYak_CAF1 14161 111 - 1 --------CGAUGGUCUGAUGAUCCGAUGCAAUUGGUGACCCACCUCCUCCUGGGAAACAGCCACCUCAUGUGGUGAACGCAUCGGGCGCGCAAAUAAAUUAGCGGCCACCAAGUGUUG --------...((((.........((((((.....((..((((........))))..)).(((((.....)))))....))))))(((.(((..........))))))))))....... ( -34.50) >consensus G_______CG__GG_C_GAUG_UCCGAUGCAAUCGGUGACCCACCUCCUCCUGGGAAACCGCCACCUCAUGUGGCGAACGCAUCGGGCGCGCAAAUAAAUUAGCGGCCACCAAGUGUUG ........................((((((....(((..((((........))))..)))(((((.....)))))....))))))(((.(((..........))))))........... (-32.60 = -32.80 + 0.20)



| Location | 13,980,379 – 13,980,498 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 70.60 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -16.18 |
| Energy contribution | -16.02 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.912858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13980379 119 - 22224390 UAUUUUUCCAGUAGUAUUACCAUCAAUGCCAUUCCAAAUCGAGCGAUUCGAUGGACCGAUGGUCCGAUGCAAUCGGUGACCCACCUCCUCCUGGGAAACCGCCACCUCAUGUGGCGAAC ...((((((((..(((((((((((....(((((...((((....)))).)))))...))))))..)))))....((((...)))).....)))))))).((((((.....))))))... ( -40.00) >DroSec_CAF1 13443 98 - 1 UAUUUUUCCAGUAGCAUUACCAUCAAUGCCAUUCCAAAUCG---------------------CCCGAUGCGAUCGGUGGCCCAGCUCCUCCUGGGAAACCGCCACCUCAUGUGGCGAAC .............(((((......)))))........((((---------------------(.....))))).(((..(((((......)))))..)))(((((.....))))).... ( -29.90) >DroSim_CAF1 13522 98 - 1 UAUUUUUCCAGUAGCAUUACCAUCAAUGCCAUUCCAAAUCG---------------------CCCGAUGCGAUCGGUGGCCCAGCUCCUCCUGGGAAACCGCCACCUCAUGUGGCGAAC .............(((((......)))))........((((---------------------(.....))))).(((..(((((......)))))..)))(((((.....))))).... ( -29.90) >DroEre_CAF1 7838 109 - 1 AAUCUUCUGAAGACGUUCAGCUUC---------CUAAGUCG-UUGAUUCGGAGGACCGAUGGUCUGAUGCCAUCGGUGACCCACCUCCUCCUGGGAAACAGCCACCUCAUGUGGUGAAC ...((((((((.....(((((..(---------....)..)-))))))))))))(((((((((.....)))))))))..((((........)))).....(((((.....))))).... ( -35.70) >DroYak_CAF1 14201 102 - 1 AAUGUCUCAAAGACCUUUAUUAUC---------UCAAGUC--------CGAUGGUCUGAUGAUCCGAUGCAAUUGGUGACCCACCUCCUCCUGGGAAACAGCCACCUCAUGUGGUGAAC ..((..(((.(((((.........---------.......--------....)))))..)))..))..((....((((...))))(((.....)))....))((((......))))... ( -20.65) >consensus UAUUUUUCCAGUAGCAUUACCAUCAAUGCCAUUCCAAAUCG_______CG__GG_C_GAUG_UCCGAUGCAAUCGGUGACCCACCUCCUCCUGGGAAACCGCCACCUCAUGUGGCGAAC ...............................................................(((((...)))))...((((........))))....((((((.....))))))... (-16.18 = -16.02 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:33 2006