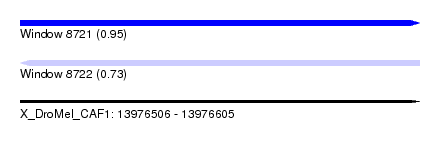
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,976,506 – 13,976,605 |
| Length | 99 |
| Max. P | 0.952691 |

| Location | 13,976,506 – 13,976,605 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.48 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -18.13 |
| Energy contribution | -18.22 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13976506 99 + 22224390 UACCGGGUGGCGUAUGACGUAUGCUGGCGAUUAAUUUCGCUCUGUGCGUAAUCAAGUCGAAAAUUUAUUGAAGUUGCAAUAAUUGCGCCUGCGGCGAA---------------------A ..(((.(.((((((((((.(((((.(((((......)))))....))))).....)))).....((((((......)))))).))))))).)))....---------------------. ( -31.00) >DroVir_CAF1 13006 106 + 1 ---------GCGUAUGACGUAUGCGAGCGAUUAAUUUCGCUCAGUGCGUAAUCAACUCGAAAAUUUAUUGAAGUUGCAAUAAUUGCGCCCAUGGCGUACCAUCAACUCAAC-----UCAA ---------(.((.(((.((((((((((((......)))))).(.(((((((((((((((.......))).))))).....))))))).)...))))))..))))).)...-----.... ( -30.90) >DroPse_CAF1 112638 99 + 1 GGCAUACCGACGUAUGACGUAUGCCGACGAUUAAUUUCGCUCUGUGCGUAAUCAAGUCAAAAAUUUAUUGAAGUUGCAAUAAAUGCGCCCAAGGCAGC---------------------G (((((((...........)))))))((((((((....(((.....))))))))..)))....((((((((......))))))))(((((...))).))---------------------. ( -26.40) >DroYak_CAF1 10305 99 + 1 UGCCGGGUGGCGUAUGACGUAUGCAGGCGAUUAAUUUCGCUCUGUGCGUAAUCAAGUCGAAAAUUUAUUGAAGUUGCAAUAAUUGCGCCUGCGGCGAA---------------------A .((((.(.((((((((((.(((((((((((......)))))...)))))).....)))).....((((((......)))))).))))))).))))...---------------------. ( -36.40) >DroMoj_CAF1 13026 120 + 1 UGCCGCAUGCCGCAUGACGUAUGCGAGCGAUUAAUUUCGCUCAGUGCGUAAUCAACUCGAAAAUUUAUUGAAGUUGCAAUAAUUGCGCCCAUGGCGUGCCAUCAGCUCAAACCCAGUCGA ....((((((((((((...)))))((((((......)))))).(.(((((((((((((((.......))).))))).....))))))).)..)))))))..................... ( -37.80) >DroPer_CAF1 133283 99 + 1 GGCAUACCGACGUAUGACGUAUGCCGACGAUUAAUUUCGCUCUGUGCGUAAUCAAGUCAAAAAUUUAUUGAAGUUGCAAUAAAUGCGCCCAAGGCAGC---------------------G (((((((...........)))))))((((((((....(((.....))))))))..)))....((((((((......))))))))(((((...))).))---------------------. ( -26.40) >consensus UGCCGACUGGCGUAUGACGUAUGCCGGCGAUUAAUUUCGCUCUGUGCGUAAUCAAGUCGAAAAUUUAUUGAAGUUGCAAUAAUUGCGCCCAAGGCGAA_____________________A ...........(((((((.(((((.(((((......)))))....))))).....)))).....((((((......)))))).)))(((...)))......................... (-18.13 = -18.22 + 0.09)

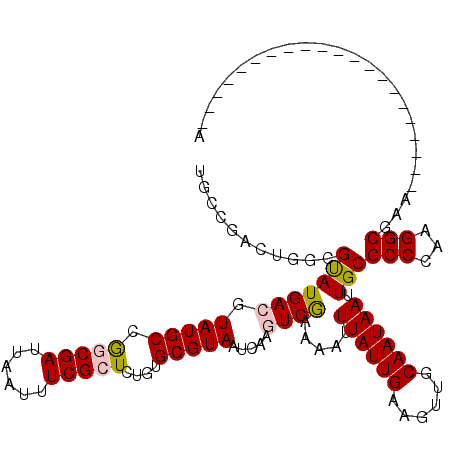

| Location | 13,976,506 – 13,976,605 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.48 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -15.72 |
| Energy contribution | -15.50 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
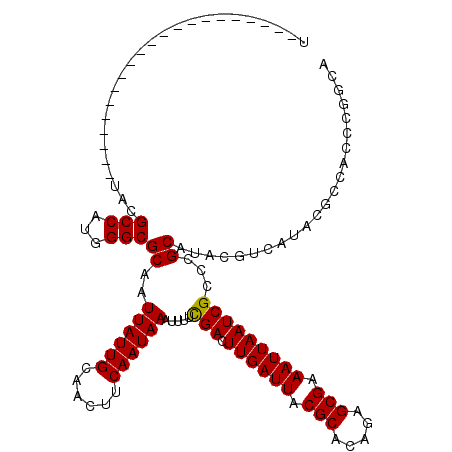
>X_DroMel_CAF1 13976506 99 - 22224390 U---------------------UUCGCCGCAGGCGCAAUUAUUGCAACUUCAAUAAAUUUUCGACUUGAUUACGCACAGAGCGAAAUUAAUCGCCAGCAUACGUCAUACGCCACCCGGUA .---------------------...((((..((((...((((((......)))))).....(((.((((((.(((.....))).)))))))))...............))))...)))). ( -22.20) >DroVir_CAF1 13006 106 - 1 UUGA-----GUUGAGUUGAUGGUACGCCAUGGGCGCAAUUAUUGCAACUUCAAUAAAUUUUCGAGUUGAUUACGCACUGAGCGAAAUUAAUCGCUCGCAUACGUCAUACGC--------- ...(-----((((.(((.((((....)))).))).)))))....((((((.((......)).))))))((.(((...(((((((......)))))))....))).))....--------- ( -27.20) >DroPse_CAF1 112638 99 - 1 C---------------------GCUGCCUUGGGCGCAUUUAUUGCAACUUCAAUAAAUUUUUGACUUGAUUACGCACAGAGCGAAAUUAAUCGUCGGCAUACGUCAUACGUCGGUAUGCC .---------------------((.(((...)))))((((((((......))))))))....(((((((((.(((.....))).))))))..)))((((((((.(....).).))))))) ( -26.00) >DroYak_CAF1 10305 99 - 1 U---------------------UUCGCCGCAGGCGCAAUUAUUGCAACUUCAAUAAAUUUUCGACUUGAUUACGCACAGAGCGAAAUUAAUCGCCUGCAUACGUCAUACGCCACCCGGCA .---------------------...((((((((((((.....))).................((.((((((.(((.....))).))))))))))))))...((.....))......))). ( -29.10) >DroMoj_CAF1 13026 120 - 1 UCGACUGGGUUUGAGCUGAUGGCACGCCAUGGGCGCAAUUAUUGCAACUUCAAUAAAUUUUCGAGUUGAUUACGCACUGAGCGAAAUUAAUCGCUCGCAUACGUCAUGCGGCAUGCGGCA ((((((.((.(((.(((.((((....)))).))).)))((((((......))))))....)).))))))...((((..((((((......))))))((((.....))))....))))... ( -40.30) >DroPer_CAF1 133283 99 - 1 C---------------------GCUGCCUUGGGCGCAUUUAUUGCAACUUCAAUAAAUUUUUGACUUGAUUACGCACAGAGCGAAAUUAAUCGUCGGCAUACGUCAUACGUCGGUAUGCC .---------------------((.(((...)))))((((((((......))))))))....(((((((((.(((.....))).))))))..)))((((((((.(....).).))))))) ( -26.00) >consensus U_____________________UACGCCAUGGGCGCAAUUAUUGCAACUUCAAUAAAUUUUCGACUUGAUUACGCACAGAGCGAAAUUAAUCGCCCGCAUACGUCAUACGCCACCCGGCA .........................(((...)))((..((((((......)))))).....(((.((((((.(((.....))).)))))))))...))...................... (-15.72 = -15.50 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:26 2006