| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,974,295 – 13,974,386 |
| Length | 91 |
| Max. P | 0.797074 |

| Location | 13,974,295 – 13,974,386 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 74.23 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -10.55 |
| Energy contribution | -11.30 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.672514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13974295 91 + 22224390 --CCCCCACCCCUCCCCCCCGCGAUUCACCCAUCGGCUUAGUUCGCAGUUUAGCGCGAAAAAAUUGAGGGG------AGGGGGGAUCAAAUAACGCAUC --...((.(((((((((....((((......))))......(((((........)))))........))))------)))))))............... ( -32.50) >DroSec_CAF1 7545 74 + 1 CCCCCCAAUC-----------UGAUUCCCCC--CGGCUUAGUUCGCAGUUUAGCGCGAA--ACUUGAGGG----------GGGGAUCAAAUAACGCAUC ..........-----------((((((((((--(..(....(((((........)))))--....).)))----------))))))))........... ( -28.70) >DroSim_CAF1 7561 75 + 1 CCCCCCCAUC-----------UGAUUCCCCC--CGGCUUAGUUCGCAGUUUAGCGCGAAAAAAUUGAGGG-----------GGGAUCAAAUAACGCAUC ..........-----------((((((((((--(((.((..(((((........)))))..))))).)))-----------)))))))........... ( -25.40) >DroYak_CAF1 7951 93 + 1 CCCCCCCAUUC--CCCCCCCUCCAUUCCCGC--CAGCUUAGUUCGCAGUUUAGCGCGAAAAAA--GAAGGGAGCGUGGGGGGGGUUUAAAUAACGCAUC ...........--(((((((..(.(((((..--...(((..(((((........)))))..))--)..))))).)..)))))))............... ( -35.80) >consensus CCCCCCCAUC___________CGAUUCCCCC__CGGCUUAGUUCGCAGUUUAGCGCGAAAAAAUUGAGGG__________GGGGAUCAAAUAACGCAUC ..(((((.....................(......).....(((((........)))))....................)))))............... (-10.55 = -11.30 + 0.75)

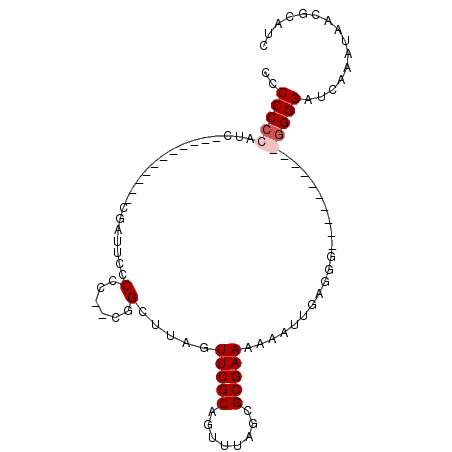
| Location | 13,974,295 – 13,974,386 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 74.23 |
| Mean single sequence MFE | -35.97 |
| Consensus MFE | -13.05 |
| Energy contribution | -15.86 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.797074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13974295 91 - 22224390 GAUGCGUUAUUUGAUCCCCCCU------CCCCUCAAUUUUUUCGCGCUAAACUGCGAACUAAGCCGAUGGGUGAAUCGCGGGGGGGAGGGGUGGGGG-- ..............((((((((------((((((......((((((......))))))....((.(((......))))).))))))))))).)))..-- ( -38.80) >DroSec_CAF1 7545 74 - 1 GAUGCGUUAUUUGAUCCCC----------CCCUCAAGU--UUCGCGCUAAACUGCGAACUAAGCCG--GGGGGAAUCA-----------GAUUGGGGGG ..(.(.(.(((((((((((----------((....(((--(.((((......)))))))).....)--))))).))))-----------))).).).). ( -26.80) >DroSim_CAF1 7561 75 - 1 GAUGCGUUAUUUGAUCCC-----------CCCUCAAUUUUUUCGCGCUAAACUGCGAACUAAGCCG--GGGGGAAUCA-----------GAUGGGGGGG ..(.(.((((((((((((-----------(((....((..((((((......))))))..))...)--))))).))))-----------))))).).). ( -30.10) >DroYak_CAF1 7951 93 - 1 GAUGCGUUAUUUAAACCCCCCCCACGCUCCCUUC--UUUUUUCGCGCUAAACUGCGAACUAAGCUG--GCGGGAAUGGAGGGGGGG--GAAUGGGGGGG ................((((((((..((((((((--((..(((.(((((....((.......))))--))).)))..)))))))))--)..)))))))) ( -48.20) >consensus GAUGCGUUAUUUGAUCCCC__________CCCUCAAUUUUUUCGCGCUAAACUGCGAACUAAGCCG__GGGGGAAUCA___________GAUGGGGGGG ....(.((((((...(((((((.......(((((......((((((......))))))..........)))))......)))))))..)))))).)... (-13.05 = -15.86 + 2.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:23 2006