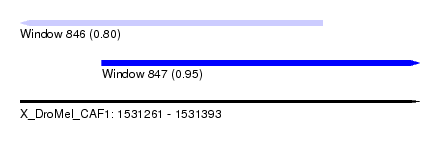
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,531,261 – 1,531,393 |
| Length | 132 |
| Max. P | 0.954443 |

| Location | 1,531,261 – 1,531,361 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 84.14 |
| Mean single sequence MFE | -15.30 |
| Consensus MFE | -14.02 |
| Energy contribution | -13.80 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
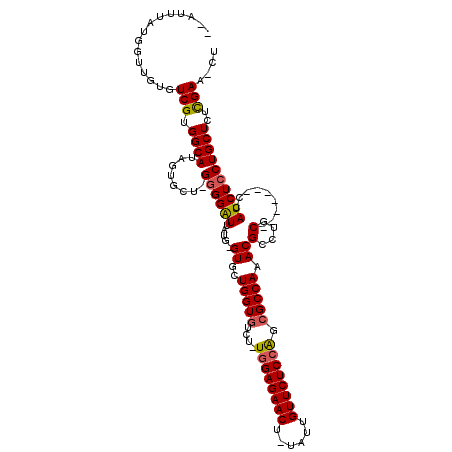
>X_DroMel_CAF1 1531261 100 - 22224390 CGUUGGAGAACAAUA-AGUUCUCCAAGACACCAGCAC-CAUAAUCCCC-AGCACUAUGCCACGACACAGCCAUUAA----------------AUCCCAGCAUCCCUACAUCCAUACAUG ..(((((((((....-.))))))))).......((..-..........-.((.....))..(......).......----------------......))................... ( -13.60) >DroSec_CAF1 4227 119 - 1 CGCUGGAGAACAAUAAAGUUCUCCAAGAAACCAGCACCCGUAAUCCUCAAGCACAAUGCCACGACACAACCAUAAAUCCCCAGUAUCCCUGUAUCCCUGCAUCCCUACAUCCAUAAAUG .((((((((((......))))))).........((....))........)))...((((...((.(((.....................))).))...))))................. ( -16.10) >DroSim_CAF1 3688 115 - 1 CGCUGGAGAACAAUA-AGUUCUCCAAGACACCAGCAC-CGUAAUCCCC-AGCACAAUGCCACGACACAACCAUAAAUC-CCAGUAUCCCUGUAUCCCUGCAUCCCUACAUCCAUACAUG .((((((((((....-.)))))))..(....))))..-.(((......-.((.....))...................-...(((....((((....))))....))).....)))... ( -16.20) >consensus CGCUGGAGAACAAUA_AGUUCUCCAAGACACCAGCAC_CGUAAUCCCC_AGCACAAUGCCACGACACAACCAUAAAUC_CCAGUAUCCCUGUAUCCCUGCAUCCCUACAUCCAUACAUG .((((((((((......)))))))).........................((.....)).......................................))................... (-14.02 = -13.80 + -0.22)

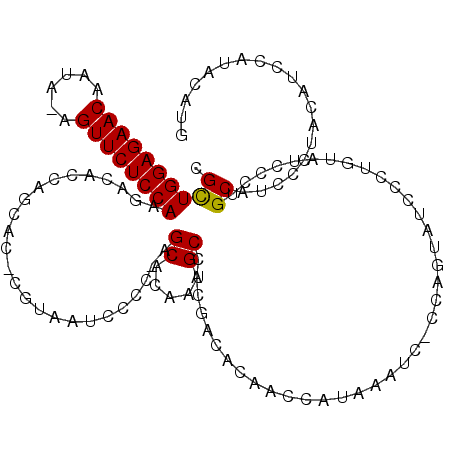
| Location | 1,531,288 – 1,531,393 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.21 |
| Mean single sequence MFE | -40.08 |
| Consensus MFE | -24.76 |
| Energy contribution | -24.88 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1531288 105 + 22224390 ---UUAAUGGCUGUGUCGUGGCAUAGUGCU-GGGGAUUAUG-GUGCUGGUGUCU-UGGAGAACU-UAUUGUUCUCCAACGCCAAACGCCUGC-------CCAUCUCCUGCUCUUGAA-CU ---......(((((((....)))))))((.-((((((...(-(((.((((((..-((((((((.-....))))))))))))))..))))...-------..)))))).)).......-.. ( -43.20) >DroSec_CAF1 4267 113 + 1 GGAUUUAUGGUUGUGUCGUGGCAUUGUGCUUGAGGAUUACGGGUGCUGGUUUCU-UGGAGAACUUUAUUGUUCUCCAGCGCCAAACGCCUGC------CCCAUCUCCUGCUCUCGAACCU ........((((((((....)))).(.((..((((....((((((.((((...(-((((((((......))))))))).))))..)))))).------....))))..)).)...)))). ( -36.30) >DroSim_CAF1 3728 107 + 1 -GAUUUAUGGUUGUGUCGUGGCAUUGUGCU-GGGGAUUACG-GUGCUGGUGUCU-UGGAGAACU-UAUUGUUCUCCAGCGCCAAACGCCUGC-------CCAUCUCCUGCUCUCGAA-CU -.......((((((((....)))).(.((.-((((((...(-(((.((((((..-((((((((.-....))))))))))))))..))))...-------..)))))).)).)...))-)) ( -37.60) >DroEre_CAF1 4221 96 + 1 ------------GUGUCGUGGCAUGGUGCU-GGGGAUUAUG-GUGAUGGUGUCU-UGGAGAACG-UAUUGUUCUCCAUCGCCAAACGCUGCC-------CCAUCUCCUGCUCUCGAC-CU ------------..((((.((((.((.(.(-((((.....(-(((.(((((...-((((((((.-....)))))))).)))))..)))).))-------))).).))))))..))))-.. ( -43.50) >DroYak_CAF1 4179 115 + 1 -AAUAUAAGGUUGUGUCGUGGCAUAGUGCU-GGGGGUUAUG-GUGCUGGUGUUUAUGGAGAACA-UGCUGUUCUCGGUCGCCAAACGGUUCCCAAACGCCCAUCUCCUGCUCUCGAC-CU -......(((((((((....))))......-((((((...(-(((..(((((((...((((((.-....))))))((..(((....)))..)))))))))))))....)))))))))-)) ( -39.80) >consensus __AUUUAUGGUUGUGUCGUGGCAUAGUGCU_GGGGAUUAUG_GUGCUGGUGUCU_UGGAGAACU_UAUUGUUCUCCAGCGCCAAACGCCUGC_______CCAUCUCCUGCUCUCGAA_CU ...............(((.((((........((((((.....((..(((((....((((((((......)))))))).))))).))(....).........))))))))))..))).... (-24.76 = -24.88 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:12 2006