
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,959,509 – 13,959,737 |
| Length | 228 |
| Max. P | 0.901615 |

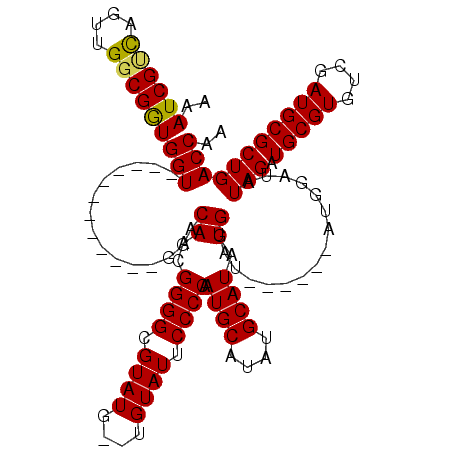
| Location | 13,959,509 – 13,959,617 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.87 |
| Mean single sequence MFE | -35.62 |
| Consensus MFE | -26.48 |
| Energy contribution | -26.23 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13959509 108 - 22224390 AAAUCGCCAGUUGGCGAUGGU------------GGACAACGGGGCGUAUGUGUGUAUUCCCCAAUGCAUAUGCAUAUGGAUAUGGAUAUGGAUAUAGAUGCGUGUCGAUGCGCUGACCAA ..((((((....))))))(((------------((.((.(((.((((((.(((((..((((((((((....)))).)))....))).....))))).)))))).))).))..)).))).. ( -38.30) >DroSec_CAF1 11008 97 - 1 -AAUCGUCAGUUGGCGGUGGU------------GGACAACGGGGCGUAU----GUAUUCCCCAAUGCAUAUGCAUAUGGAU------AUGGAUAUAGAUGCGUGUCGAUGCGCUGACCAA -.((((((....))))))(((------------((.((.(((.((((((----.(((((((((((((....)))).)))..------..))).))).)))))).))).))..)).))).. ( -33.00) >DroSim_CAF1 11655 100 - 1 AAAUCGUCAGUUGGCGGUGGU------------GGACAACGGGGCGUAUG--UGUAUUCCCCAAUGCAUAUGCAUAUGGAU------AUGGAUAUAGAUGCGUGUCGAUGCGCUGACCAA ..((((((....))))))(((------------((.((.(((.((((((.--(((((((.(((((((....)))).)))..------..))))))).)))))).))).))..)).))).. ( -35.70) >DroYak_CAF1 11463 117 - 1 AAAUCGGUUGUUGGCGGUGGUUUGGUGGUGGUGGGACAACGGGGCGUAUG--UGUAUUCCCCAAUGCAUAUGCAUAUGGAUAUGG-UAUGGAUAUAGAUGCGUGUCGAUGCGCUGACCAA ..........(((((((((((((.(......).))))..(((.((((((.--(((((((.(((((.((((....)))).)).)))-...))))))).)))))).)))...))))).)))) ( -35.50) >consensus AAAUCGUCAGUUGGCGGUGGU____________GGACAACGGGGCGUAUG__UGUAUUCCCCAAUGCAUAUGCAUAUGGAU______AUGGAUAUAGAUGCGUGUCGAUGCGCUGACCAA ..((((((....))))))(((...............((..((((.((((....)))).)))).((((....)))).))................(((.(((((....))))))))))).. (-26.48 = -26.23 + -0.25)

| Location | 13,959,549 – 13,959,657 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.89 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -25.30 |
| Energy contribution | -25.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13959549 108 - 22224390 GUUGCCUUUGUAAUUAAAAAGGAAAAGGAAAAUACGACAAAAAUCGCCAGUUGGCGAUGGU------------GGACAACGGGGCGUAUGUGUGUAUUCCCCAAUGCAUAUGCAUAUGGA (((((((((........)))))............(.((....((((((....)))))).))------------.).))))((((.((((....)))).)))).((((....))))..... ( -32.60) >DroSec_CAF1 11042 93 - 1 GUUGCCUUUGUAAUUAAAAAGGAAAAGGAA-----------AAUCGUCAGUUGGCGGUGGU------------GGACAACGGGGCGUAU----GUAUUCCCCAAUGCAUAUGCAUAUGGA (((((((((........)))))........-----------.((((((....))))))...------------...))))...((((((----(((((....)))))))))))....... ( -25.90) >DroSim_CAF1 11689 106 - 1 GUUGCCUUUGUAAUUAAAAAGGAAAAGGAAAAUACGACAAAAAUCGUCAGUUGGCGGUGGU------------GGACAACGGGGCGUAUG--UGUAUUCCCCAAUGCAUAUGCAUAUGGA (((((((((........)))))............(.((....((((((....)))))).))------------.).))))((((.(((..--..))).)))).((((....))))..... ( -27.80) >DroYak_CAF1 11502 112 - 1 GUUUCCUUUGUAAUUAAAAAGGAA------AAUACGACAAAAAUCGGUUGUUGGCGGUGGUUUGGUGGUGGUGGGACAACGGGGCGUAUG--UGUAUUCCCCAAUGCAUAUGCAUAUGGA .((((((((........)))))))------)...((((((.......)))))).(.(((((((.((.((......)).)).))))(((((--((((((....))))))))))).))).). ( -28.40) >consensus GUUGCCUUUGUAAUUAAAAAGGAAAAGGAAAAUACGACAAAAAUCGUCAGUUGGCGGUGGU____________GGACAACGGGGCGUAUG__UGUAUUCCCCAAUGCAUAUGCAUAUGGA (((((((((........)))))....................((((((....))))))..................))))((((.((((....)))).)))).((((....))))..... (-25.30 = -25.30 + 0.00)


| Location | 13,959,589 – 13,959,697 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -22.11 |
| Energy contribution | -22.42 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13959589 108 - 22224390 GAGGGAUUUGGAAGGCGGCAAAUAGCGCUGCAUCCUUCUUGUUGCCUUUGUAAUUAAAAAGGAAAAGGAAAAUACGACAAAAAUCGCCAGUUGGCGAUGGU------------GGACAAC (((((((.......(((((.......))))))))))))(((((.(((((........)))))...............((...((((((....))))))..)------------)))))). ( -32.01) >DroSec_CAF1 11078 97 - 1 GAGGGAAUCGGUGGGCGGAAAAGAGCGCUGCAUCCUUCUUGUUGCCUUUGUAAUUAAAAAGGAAAAGGAA-----------AAUCGUCAGUUGGCGGUGGU------------GGACAAC ((((((..(((((..(......)..)))))..))))))(((((.(((((........)))))........-----------.((((((....))))))...------------.))))). ( -26.80) >DroSim_CAF1 11727 108 - 1 GAGGGAAUCGGUGGGCGGAAGAGAGCGCUGCAUCCUUCUUGUUGCCUUUGUAAUUAAAAAGGAAAAGGAAAAUACGACAAAAAUCGUCAGUUGGCGGUGGU------------GGACAAC ((((((..(((((..(....)....)))))..))))))(((((.(((((........)))))...............((...((((((....))))))..)------------)))))). ( -29.40) >DroYak_CAF1 11540 110 - 1 GAGGGAAUCG----GCGAAAAAGAGCGCUGCAUCCUUCUUGUUUCCUUUGUAAUUAAAAAGGAA------AAUACGACAAAAAUCGGUUGUUGGCGGUGGUUUGGUGGUGGUGGGACAAC ((((((..((----(((........)))))..))))))(((((((((((........)))))).------..(((.(((((.((((.(....).))))..))).)).)))....))))). ( -29.10) >consensus GAGGGAAUCGGUGGGCGGAAAAGAGCGCUGCAUCCUUCUUGUUGCCUUUGUAAUUAAAAAGGAAAAGGAAAAUACGACAAAAAUCGUCAGUUGGCGGUGGU____________GGACAAC ((((((..((((..((........))))))..))))))(((((.(((((........)))))....................((((((....))))))................))))). (-22.11 = -22.42 + 0.31)

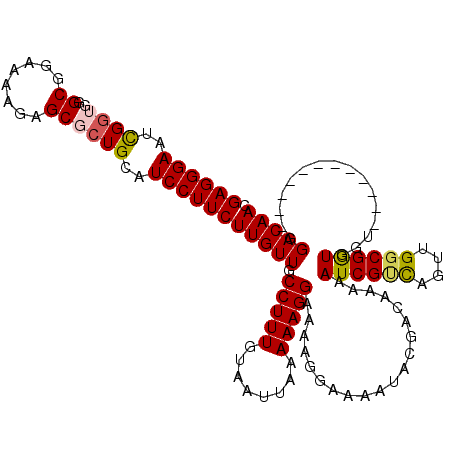
| Location | 13,959,617 – 13,959,737 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.86 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -20.99 |
| Energy contribution | -21.55 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13959617 120 - 22224390 GCCGCCCACCGGAAAAUAUAUGUGCACAAAUAGCGAGCGAGAGGGAUUUGGAAGGCGGCAAAUAGCGCUGCAUCCUUCUUGUUGCCUUUGUAAUUAAAAAGGAAAAGGAAAAUACGACAA .(((.....)))........(((((.......)).((((((((((((.......(((((.......))))))))))))))))).(((((........)))))..............))). ( -28.81) >DroSec_CAF1 11105 110 - 1 GCUACCCACCGGAAAAUAUAUGUGGAGAAAUAGCGAGCGAGAGGGAAUCGGUGGGCGGAAAAGAGCGCUGCAUCCUUCUUGUUGCCUUUGUAAUUAAAAAGGAAAAGGAA---------- (((((((((............)))).)...)))).(((((((((((..(((((..(......)..)))))..))))))))))).(((((........)))))........---------- ( -31.50) >DroSim_CAF1 11755 120 - 1 GCUGCCCACCGGAAAAUAUAUGUGCACAAAUAGCGAGCGAGAGGGAAUCGGUGGGCGGAAGAGAGCGCUGCAUCCUUCUUGUUGCCUUUGUAAUUAAAAAGGAAAAGGAAAAUACGACAA .(((((((((((..........(((.......)))..(....)....))))))))))).................(((((....(((((........)))))...))))).......... ( -31.50) >DroYak_CAF1 11580 110 - 1 ACCUCCCACCGGAAAAUAUAUGUGCACAAAUUGAGAGCGAGAGGGAAUCG----GCGAAAAAGAGCGCUGCAUCCUUCUUGUUUCCUUUGUAAUUAAAAAGGAA------AAUACGACAA ...(((....))).......((((............((((((((((..((----(((........)))))..))))))))))(((((((........)))))))------..)))).... ( -29.60) >consensus GCCGCCCACCGGAAAAUAUAUGUGCACAAAUAGCGAGCGAGAGGGAAUCGGUGGGCGGAAAAGAGCGCUGCAUCCUUCUUGUUGCCUUUGUAAUUAAAAAGGAAAAGGAAAAUACGACAA ...................................(((((((((((..((((..((........))))))..))))))))))).(((((........))))).................. (-20.99 = -21.55 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:13 2006