| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,956,932 – 13,957,049 |
| Length | 117 |
| Max. P | 0.710926 |

| Location | 13,956,932 – 13,957,049 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.01 |
| Mean single sequence MFE | -34.41 |
| Consensus MFE | -26.71 |
| Energy contribution | -27.40 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13956932 117 + 22224390 AGAAGACCCAGCCUCCAGUGGGUUCUAUCCACGGUGGGGCAAUAAAAAUUUAUGAAGCCUGAGGAUGCUGGCGGGGGCAGGGUCUCAGCUGCUUAACCCAUUAAUGACCAAUGAG---AA .((.(((((.((((((.((.(((...((((....(.((((.((((....))))...)))).)))))))).)))))))).))))))).............................---.. ( -35.80) >DroSec_CAF1 8151 103 + 1 AGAAGACCCAGCCUCC----------------GGUGGGGCAAUAAAAAUUUAUGAAGCCAGAGGAAGCUAA-GGGGGCAGGGUCUCAGCAGCUUAACCCAUUAAAUACCAAUGAAUGUAA .((.(((((.((((((----------------.....(((.((((....))))...)))..((....))..-)))))).))))))).................................. ( -27.70) >DroSim_CAF1 8540 119 + 1 AGAAGACCCAGCCUCCAGUGGGUUUUAUCCACGGUGGGGCAAUAAAAAUUUAUGAAGCCAGAGGAAGCUGA-GGGGGCAGGGUCUCAGCAGCUUAACCCAUUAAAUACCAAUGACUGUAA .((.(((((.((((((.((((((...)))))).....(((.((((....))))...)))............-)))))).))))))).(((((....................).)))).. ( -35.45) >DroYak_CAF1 8912 116 + 1 AGAAGACCCAGCCUCCAGUGGGUUUUAUCCACAGUGGGGCAAUAAAAAUUUAUGAAGCCCGAAGAAACUGU-GGGGGCAGGGCUCC---AGCUUAACCCAUUAAAGGCCAGUGCCUGUAA ..((((((((........))))))))..(((((((.((((.((((....))))...))))......)))))-))(((..((((...---.))))..))).....((((....)))).... ( -38.70) >consensus AGAAGACCCAGCCUCCAGUGGGUUUUAUCCACGGUGGGGCAAUAAAAAUUUAUGAAGCCAGAGGAAGCUGA_GGGGGCAGGGUCUCAGCAGCUUAACCCAUUAAAGACCAAUGACUGUAA ...((((((.((((((.((((((...)))))).....(((.((((....))))...))).............)))))).))))))................................... (-26.71 = -27.40 + 0.69)

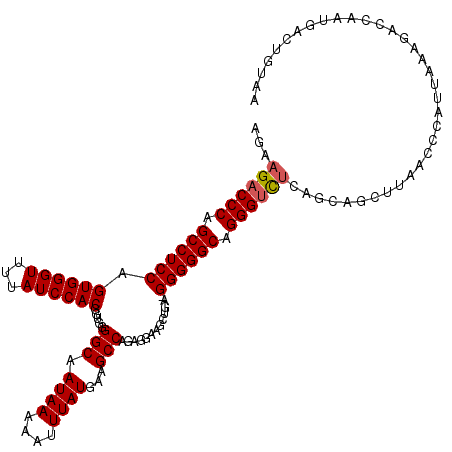

| Location | 13,956,932 – 13,957,049 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.01 |
| Mean single sequence MFE | -33.25 |
| Consensus MFE | -23.44 |
| Energy contribution | -23.50 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.710926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13956932 117 - 22224390 UU---CUCAUUGGUCAUUAAUGGGUUAAGCAGCUGAGACCCUGCCCCCGCCAGCAUCCUCAGGCUUCAUAAAUUUUUAUUGCCCCACCGUGGAUAGAACCCACUGGAGGCUGGGUCUUCU ..---(((((((.....)))))))..........(((((((.(((.(((((..........)))........................((((.......)))).)).))).))))))).. ( -32.50) >DroSec_CAF1 8151 103 - 1 UUACAUUCAUUGGUAUUUAAUGGGUUAAGCUGCUGAGACCCUGCCCCC-UUAGCUUCCUCUGGCUUCAUAAAUUUUUAUUGCCCCACC----------------GGAGGCUGGGUCUUCU ....((((((((.....)))))))).........(((((((.......-..(((((((...(((...((((....)))).))).....----------------)))))))))))))).. ( -24.50) >DroSim_CAF1 8540 119 - 1 UUACAGUCAUUGGUAUUUAAUGGGUUAAGCUGCUGAGACCCUGCCCCC-UCAGCUUCCUCUGGCUUCAUAAAUUUUUAUUGCCCCACCGUGGAUAAAACCCACUGGAGGCUGGGUCUUCU .....(((.(..((((((((....))))).)))..))))........(-(((((((((...(((...((((....)))).))).....((((.......)))).))))))))))...... ( -31.80) >DroYak_CAF1 8912 116 - 1 UUACAGGCACUGGCCUUUAAUGGGUUAAGCU---GGAGCCCUGCCCCC-ACAGUUUCUUCGGGCUUCAUAAAUUUUUAUUGCCCCACUGUGGAUAAAACCCACUGGAGGCUGGGUCUUCU ....((((.(..(((((((.((((((..((.---((...)).))..((-(((((......((((...((((....)))).)))).)))))))....)))))).)))))))..)))))... ( -44.20) >consensus UUACAGUCAUUGGUAUUUAAUGGGUUAAGCUGCUGAGACCCUGCCCCC_UCAGCUUCCUCUGGCUUCAUAAAUUUUUAUUGCCCCACCGUGGAUAAAACCCACUGGAGGCUGGGUCUUCU ...........((((......(((((..........)))))))))(((...(((((((...(((...((((....)))).))).....((((.......)))).))))))))))...... (-23.44 = -23.50 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:07 2006