
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,953,822 – 13,953,964 |
| Length | 142 |
| Max. P | 0.983984 |

| Location | 13,953,822 – 13,953,927 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 94.47 |
| Mean single sequence MFE | -25.56 |
| Consensus MFE | -22.31 |
| Energy contribution | -22.88 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13953822 105 - 22224390 GGUUGGAAAAUGUUAUCGCAACCUGUGGGCGAAGAAUCUUGAAAGAUGAUUUGCUAGACCAGCAGGGUGAUAA-AAUCAAGCACAAAUUAAGCAAAAGAUUAGAUA (.((((......(((((((..(((((.((((((..((((....))))..))))))......))))))))))))-..)))).)........................ ( -24.40) >DroSec_CAF1 5262 105 - 1 GGUUGGAAAAUGCUAUCGCAACCUGUGGGCGAAGAAUCUCGAAAGAUGAUUUGCUCGACCAGCAGGGUGAUAA-AAUCAAGCACAAAUUAGGCAAAAGAUUAGAUA .(((......(((((((((..(((((.(((((.(((((((....)).)))))..))).)).))))))))))..-.....)))).......)))............. ( -28.23) >DroSim_CAF1 5664 105 - 1 GGUUGGAAAACGCUAUCGCAACCUGUGGGCGAAGAAUCUCGAAAGAUGAUUUGCUCGACCAGCAGGGUGAUAA-AAUCAAGCACAAAUUAAGCAAAAGAUUAGAUA .(((....)))..((((((..(((((.(((((.(((((((....)).)))))..))).)).))))))))))).-......((.........))............. ( -29.30) >DroYak_CAF1 5703 106 - 1 GGAAGGAAAAUGCUAUCGCAACCUGUGGGCGAAGAAUCUCGAAAGAUGAUUUGCUGGACCAGCUGGGUGAUAAAAAUGAAGCACAAAUUUAGCAAAAGAUUAGAUA ..........(((((((((..((.((.(((.(.(((((((....)).)))))..).).)).)).))))))))....((((.......)))))))............ ( -20.30) >consensus GGUUGGAAAAUGCUAUCGCAACCUGUGGGCGAAGAAUCUCGAAAGAUGAUUUGCUCGACCAGCAGGGUGAUAA_AAUCAAGCACAAAUUAAGCAAAAGAUUAGAUA .............((((((..(((((.(((((.(((((((....)).)))))..))).)).)))))))))))........((.........))............. (-22.31 = -22.88 + 0.56)

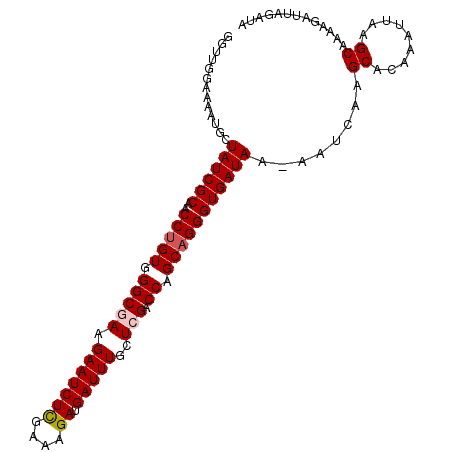
| Location | 13,953,855 – 13,953,964 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 86.38 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -22.17 |
| Energy contribution | -22.80 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13953855 109 - 22224390 GUAAUGUGAAAAAUCCAGAA---AAAAAGGAUUGAAAAGUGGUUGGAAAAUGUUAUCGCAACCUGUGGGCGAAGAAUCUUGAAAGAUGAUUUGCUAGACCAGCAGGGUGAUA ....(((((..(((((....---.....)))))........(((....)))....))))).(((((.((((((..((((....))))..))))))......)))))...... ( -25.30) >DroSec_CAF1 5295 107 - 1 GAAAUGUGAAAAAUCAGGAA-----AAAGCUUUGAAAAGCGGUUGGAAAAUGCUAUCGCAACCUGUGGGCGAAGAAUCUCGAAAGAUGAUUUGCUCGACCAGCAGGGUGAUA ....(((((....(((((..-----.....)))))..((((.((....)))))).))))).(((((.(((((.(((((((....)).)))))..))).)).)))))...... ( -31.30) >DroSim_CAF1 5697 107 - 1 GAAAUGUGAAAAAUCAGGAA-----AAAGCAUUGAAAAGCGGUUGGAAAACGCUAUCGCAACCUGUGGGCGAAGAAUCUCGAAAGAUGAUUUGCUCGACCAGCAGGGUGAUA ....(((((....((((...-----......))))..((((.((....)))))).))))).(((((.(((((.(((((((....)).)))))..))).)).)))))...... ( -32.50) >DroYak_CAF1 5737 109 - 1 ---UUGUGAAAAAUCAGGCAAAAAAAAAUGUCUAGAGAGUGGAAGGAAAAUGCUAUCGCAACCUGUGGGCGAAGAAUCUCGAAAGAUGAUUUGCUGGACCAGCUGGGUGAUA ---.........(((((((.........((((((.((..((((.((......)).)).))..)).))))))..(((((((....)).))))))))...((....)).)))). ( -24.20) >consensus GAAAUGUGAAAAAUCAGGAA_____AAAGCAUUGAAAAGCGGUUGGAAAAUGCUAUCGCAACCUGUGGGCGAAGAAUCUCGAAAGAUGAUUUGCUCGACCAGCAGGGUGAUA ............((((......................(((((.((......)))))))..(((((.(((((.(((((((....)).)))))..))).)).))))).)))). (-22.17 = -22.80 + 0.63)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:03 2006