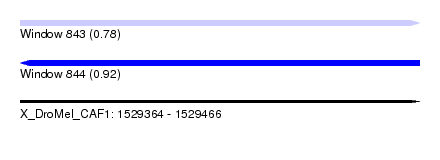
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,529,364 – 1,529,466 |
| Length | 102 |
| Max. P | 0.921543 |

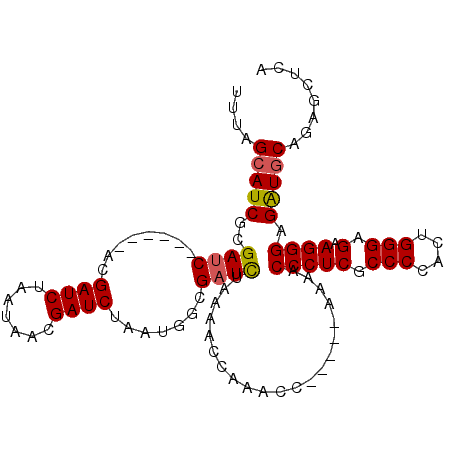
| Location | 1,529,364 – 1,529,466 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 89.76 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -25.72 |
| Energy contribution | -26.24 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.783971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1529364 102 + 22224390 UGAGCUCUGCAUCUCCCUUCUCCCAGUGGGGCGAGGGUUUU-----GGUUUGGUUUUAGAUCGCCAUUAGAUCGUUAUUAGAUCGUGAUCGUGAUCGCGAUGCUAAA ..(((..(((.....(((((.(((....))).)))))....-----............((((((.((((((((.......)))).)))).)))))))))..)))... ( -33.50) >DroSec_CAF1 2351 96 + 1 UGAGCUCUGCAUCUCCCUUCUCCCAGUGGGGCGAGGGUUUU-----GGUUUGGUUUUAGAUCGCCAUUAGAUCGUUAUUAGAUCGU------GAUCGCGAUGCUAAA .(((((..((....(((((.((((...)))).)))))....-----.))..)))))((((((((.((((((((.......)))).)------))).))))).))).. ( -31.40) >DroSim_CAF1 1806 102 + 1 UGAGCUCUGCAUCUCCCUUCUCCCAGUGGGGCGAGGGUUUU-----GGUUUGGUUUUAGAUCGCCAUUAGAUCGUUAUUAGAUCGUGAUCGUGAUCGCGAUGCUAAA ..(((..(((.....(((((.(((....))).)))))....-----............((((((.((((((((.......)))).)))).)))))))))..)))... ( -33.50) >DroEre_CAF1 2433 101 + 1 UGAUUUCUGCACCUCCCUUCUCCCAGUGGGGCGAGGGUUUUGGUUUGGUUUGGUUUUAAAUCGUCAUUAGAUCGAUACUAGAUCGU------GUUCGCGAUGCCAAA ......(((.(((.(((((.((((...)))).)))))....))).)))((((((.....(((((.....((((.......))))..------....))))))))))) ( -27.40) >DroYak_CAF1 2319 97 + 1 UGAGCUCUGCACCUCCCUUCUCCCAGUGGGGCGAGGGUUUU----CGGUUUGGUUUUAGAUCGUCAUUAGAUCGAUAUUAGAUCGU------GAUCGCGAUUCUAAA .(((((..((....(((((.((((...)))).)))))....----..))..)))))((((((((.((((((((.......)))).)------))).)))).)))).. ( -29.30) >consensus UGAGCUCUGCAUCUCCCUUCUCCCAGUGGGGCGAGGGUUUU_____GGUUUGGUUUUAGAUCGCCAUUAGAUCGUUAUUAGAUCGU______GAUCGCGAUGCUAAA ..(((..(((.....(((((.(((....))).))))).........((((((((....((((.......))))...))))))))............)))..)))... (-25.72 = -26.24 + 0.52)

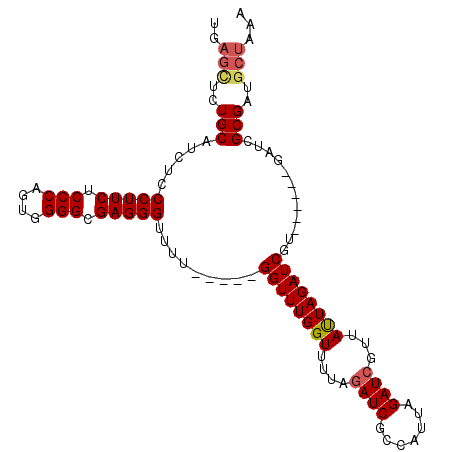
| Location | 1,529,364 – 1,529,466 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 89.76 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -21.74 |
| Energy contribution | -21.74 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1529364 102 - 22224390 UUUAGCAUCGCGAUCACGAUCACGAUCUAAUAACGAUCUAAUGGCGAUCUAAAACCAAACC-----AAAACCCUCGCCCCACUGGGAGAAGGGAGAUGCAGAGCUCA ....(((((..((((.(.((...((((.......))))..)).).))))............-----....(((((.(((....))).).)))).)))))........ ( -27.30) >DroSec_CAF1 2351 96 - 1 UUUAGCAUCGCGAUC------ACGAUCUAAUAACGAUCUAAUGGCGAUCUAAAACCAAACC-----AAAACCCUCGCCCCACUGGGAGAAGGGAGAUGCAGAGCUCA (((((.(((((.((.------..((((.......))))..)).))))))))))........-----....(((((.(((....))).).)))).((.((...)))). ( -25.00) >DroSim_CAF1 1806 102 - 1 UUUAGCAUCGCGAUCACGAUCACGAUCUAAUAACGAUCUAAUGGCGAUCUAAAACCAAACC-----AAAACCCUCGCCCCACUGGGAGAAGGGAGAUGCAGAGCUCA ....(((((..((((.(.((...((((.......))))..)).).))))............-----....(((((.(((....))).).)))).)))))........ ( -27.30) >DroEre_CAF1 2433 101 - 1 UUUGGCAUCGCGAAC------ACGAUCUAGUAUCGAUCUAAUGACGAUUUAAAACCAAACCAAACCAAAACCCUCGCCCCACUGGGAGAAGGGAGGUGCAGAAAUCA ....(((((......------..((((.......))))................................(((((.(((....))).).)))).)))))........ ( -21.10) >DroYak_CAF1 2319 97 - 1 UUUAGAAUCGCGAUC------ACGAUCUAAUAUCGAUCUAAUGACGAUCUAAAACCAAACCG----AAAACCCUCGCCCCACUGGGAGAAGGGAGGUGCAGAGCUCA ....((.((((((((------.((((.....)))).((....)).)))).........(((.----....(((((.(((....))).).)))).))))).))..)). ( -21.80) >consensus UUUAGCAUCGCGAUC______ACGAUCUAAUAACGAUCUAAUGGCGAUCUAAAACCAAACC_____AAAACCCUCGCCCCACUGGGAGAAGGGAGAUGCAGAGCUCA ....(((((..((((........((((.......)))).......)))).....................(((((.(((....))).).)))).)))))........ (-21.74 = -21.74 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:10 2006