
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,933,563 – 13,933,760 |
| Length | 197 |
| Max. P | 0.769550 |

| Location | 13,933,563 – 13,933,680 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.72 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -22.16 |
| Energy contribution | -22.24 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13933563 117 + 22224390 CGUGGGUCAAUGACUUGGCCAAAA-GUUUUUUUUUUGGUUAUUGCUUUUGUUAUUACAACAGCCCACACAC--AUGCCCUGUCAUCCGUGGGCCACUUUUUCUUCUGUCAUUCUUAUUUC .((((.((((((((.(((((((((-(.....))))))))))..(((.((((....)))).)))........--.......)))))...))).))))........................ ( -27.40) >DroSec_CAF1 13466 116 + 1 CGUGGGUCAAUGACUUGGCCAAAA-GCUUUU-GUUUGGUUAUUGCUUUUGUUAUUACAACAGCCCACACAC--AUGCCCUGUCAUCCGUGGGCCACUUUUUCUUCUGCCAUUCUUAUUUC .((((.((((((((..(((...((-((....-))))((.....(((.((((....)))).)))))......--..)))..)))))...))).))))........................ ( -25.80) >DroSim_CAF1 18266 116 + 1 CGUGGGUCAAUGACUUGGCCAAAA-GCUUUU-GUUUGGUUAUUGCUUUUGUUAUUACAACAGCCCACACAC--AUGCCCUGUCAUCCGUGGGCCACUUUUUCUUCUGCCAUUCUUAUUUC .((((.((((((((..(((...((-((....-))))((.....(((.((((....)))).)))))......--..)))..)))))...))).))))........................ ( -25.80) >DroEre_CAF1 14999 117 + 1 CGUGGGUCAACGACUUGGCCAAAA-GCUUUU-GUUUGGUUAUUGCUUUCGGUACUACAACAGCCCACACAU-CCUGCCAUGUCAUCCGUGGGCCACUUUUUCUUCUGCCAUUCUUAUUUC .(((((((((....))))))....-.....(-(((((((.((((....)))))))).))))((((((((((-......)))).....)))))))))........................ ( -23.70) >DroYak_CAF1 16421 119 + 1 CGUGGGUCAGUGACUUGGCCAGAAUGCUUUU-GUUUGGUUAUUGCUUUUGUUAUUACAACAGCCCACUCACAUGUGCCCUGUCAUCCGUGGGCCACUUUUUCUUCUGCCAUUCUUAUUUC .((((.((((((((..(((....(((....(-(..(((.....(((.((((....)))).))))))..)))))..)))..)))))...))).))))........................ ( -27.20) >consensus CGUGGGUCAAUGACUUGGCCAAAA_GCUUUU_GUUUGGUUAUUGCUUUUGUUAUUACAACAGCCCACACAC__AUGCCCUGUCAUCCGUGGGCCACUUUUUCUUCUGCCAUUCUUAUUUC .((((.((((((((.((((((((..........))))))))..(((.(((......))).))).................)))))...))).))))........................ (-22.16 = -22.24 + 0.08)

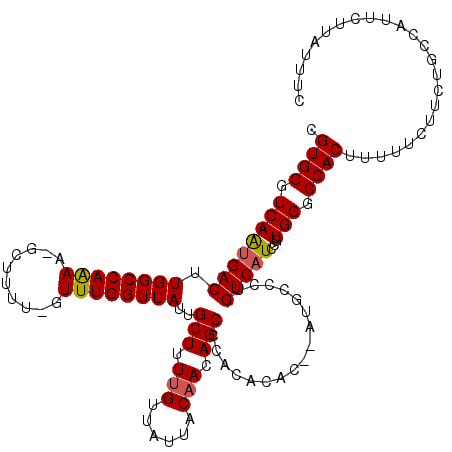
| Location | 13,933,602 – 13,933,720 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.10 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -25.47 |
| Energy contribution | -25.49 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.769550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13933602 118 - 22224390 UCUGUCGGCCUUAUCAAGUCGAGGGAAAAAAAGCCGAGCCGAAAUAAGAAUGACAGAAGAAAAAGUGGCCCACGGAUGACAGGGCAU--GUGUGUGGGCUGUUGUAAUAACAAAAGCAAU (((((((..(((((....(((.((.........))....))).)))))..)))))))......((..(((((((.((..........--)).)))))))..))................. ( -30.90) >DroSec_CAF1 13504 117 - 1 UCUGUCGGCCUUAUCAAGUCGAGGUA-AAAAAGCCGAGCCGAAAUAAGAAUGGCAGAAGAAAAAGUGGCCCACGGAUGACAGGGCAU--GUGUGUGGGCUGUUGUAAUAACAAAAGCAAU (((((((..(((((....(((.(((.-.....)))....))).)))))..)))))))......((..(((((((.((..........--)).)))))))..))................. ( -32.90) >DroSim_CAF1 18304 117 - 1 UCUGUCGGCCUUAUCAAGUCGAGGUA-AAAAAGCCGAGCCGAAAUAAGAAUGGCAGAAGAAAAAGUGGCCCACGGAUGACAGGGCAU--GUGUGUGGGCUGUUGUAAUAACAAAAGCAAU (((((((..(((((....(((.(((.-.....)))....))).)))))..)))))))......((..(((((((.((..........--)).)))))))..))................. ( -32.90) >DroEre_CAF1 15037 99 - 1 UCUAUCAGCCUUAUCAA--------------------GCCGAAAUAAGAAUGGCAGAAGAAAAAGUGGCCCACGGAUGACAUGGCAGG-AUGUGUGGGCUGUUGUAGUACCGAAAGCAAU (((..((..(((((...--------------------......)))))..))..)))......((..(((((((.((..(......).-)).)))))))..)).......(....).... ( -22.30) >DroYak_CAF1 16460 119 - 1 UCUGUCGGCCUUAUCAAGUUGAGGGA-AAAAAGCCGAGCCGAAAUAAGAAUGGCAGAAGAAAAAGUGGCCCACGGAUGACAGGGCACAUGUGAGUGGGCUGUUGUAAUAACAAAAGCAAU (((((((..(((((...(((..((..-......)).)))....)))))..)))))))......((..((((((...(.(((.......))).)))))))..))................. ( -29.00) >consensus UCUGUCGGCCUUAUCAAGUCGAGGGA_AAAAAGCCGAGCCGAAAUAAGAAUGGCAGAAGAAAAAGUGGCCCACGGAUGACAGGGCAU__GUGUGUGGGCUGUUGUAAUAACAAAAGCAAU (((((((..(((((........((.........))........)))))..)))))))......((..(((((((.((............)).)))))))..))................. (-25.47 = -25.49 + 0.02)



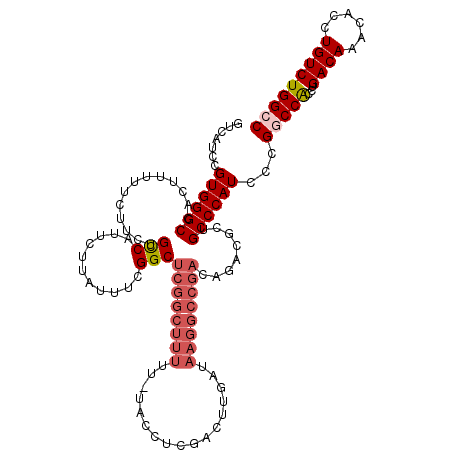
| Location | 13,933,640 – 13,933,760 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.45 |
| Mean single sequence MFE | -30.64 |
| Consensus MFE | -23.37 |
| Energy contribution | -25.05 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13933640 120 + 22224390 GUCAUCCGUGGGCCACUUUUUCUUCUGUCAUUCUUAUUUCGGCUCGGCUUUUUUUCCCUCGACUUGAUAAGGCCGACAGACGCUGCCCAUACCGGCCACCUGACAAACACCUGUCUGGCC .......((((((..........((((((..(((((((..((.((((...........)))))).)))))))..))))))....))))))...(((((...((((......))))))))) ( -32.44) >DroSec_CAF1 13542 119 + 1 GUCAUCCGUGGGCCACUUUUUCUUCUGCCAUUCUUAUUUCGGCUCGGCUUUUU-UACCUCGACUUGAUAAGGCCGACAGACGCUGCCCAUCCCGGCCACCUGACAAACACCUGUCUGGCC .......((((((.............(((...........)))((((((((.(-((........))).))))))))........))))))...(((((...((((......))))))))) ( -33.50) >DroSim_CAF1 18342 119 + 1 GUCAUCCGUGGGCCACUUUUUCUUCUGCCAUUCUUAUUUCGGCUCGGCUUUUU-UACCUCGACUUGAUAAGGCCGACAGACGCUGCCCAUCCCGGCCACCUGACAAACACCUGUCUGGCC .......((((((.............(((...........)))((((((((.(-((........))).))))))))........))))))...(((((...((((......))))))))) ( -33.50) >DroEre_CAF1 15076 100 + 1 GUCAUCCGUGGGCCACUUUUUCUUCUGCCAUUCUUAUUUCGGC--------------------UUGAUAAGGCUGAUAGACGCUGCCCAUCCCGCCCGCCUGACAAACACCUGUCUGGCC .......((((((.............((...(((....(((((--------------------((....))))))).))).)).)))))).......(((.((((......)))).))). ( -26.44) >DroYak_CAF1 16500 119 + 1 GUCAUCCGUGGGCCACUUUUUCUUCUGCCAUUCUUAUUUCGGCUCGGCUUUUU-UCCCUCAACUUGAUAAGGCCGACAGACGCUGCCCAUCCCGUCCACCUGACAAACACCUGUCUGGCC .......((((((.............(((...........)))((((((((..-((.........)).))))))))........))))))........((.((((......)))).)).. ( -27.30) >consensus GUCAUCCGUGGGCCACUUUUUCUUCUGCCAUUCUUAUUUCGGCUCGGCUUUUU_UACCUCGACUUGAUAAGGCCGACAGACGCUGCCCAUCCCGGCCACCUGACAAACACCUGUCUGGCC .......((((((.............(((...........)))((((((((.................))))))))........))))))...(((((...((((......))))))))) (-23.37 = -25.05 + 1.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:47 2006