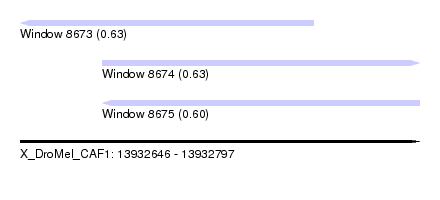
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,932,646 – 13,932,797 |
| Length | 151 |
| Max. P | 0.632758 |

| Location | 13,932,646 – 13,932,757 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.11 |
| Mean single sequence MFE | -28.96 |
| Consensus MFE | -24.20 |
| Energy contribution | -24.60 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13932646 111 - 22224390 GUAUCUUAUGUGAUGAAAUUUAUGCUUAAUUAAAAGCGACACAAGGCGUCACAAGCAACGCACAACGCGUUCGCCCAGAUAAGAUGCGAA---------AGAACCAAGCGGGCCAUCGGC (((((((((((((((.......(((((......)))))........))))))..(((((((.....))))).))....)))))))))...---------....((....))(((...))) ( -30.66) >DroSec_CAF1 12601 111 - 1 GUAUCUUAUGUGAUGAAAUUUAUGCUUAAUUAAAAGCGACACAAGGCGUCACAAGCAACGCACAACGCGUUCGCUCAGAUAAGACGCGAA---------AUAACCAAGCGGGCCAUCGGC (..((((((((((((.......(((((......)))))........)))))).((((((((.....))))).)))...))))))..)...---------....((....))(((...))) ( -28.36) >DroSim_CAF1 16165 111 - 1 GUAUCUUAUGUGAUGAAAUUUAUGCUUAAUUAAAAGCGACACAAGGCGUCACAAGCAACGCACAACGCGUUCGCCCAGAUAAGACGCGAA---------AUAACCGAGCGGGCCAUCGGC (..((((((((((((.......(((((......)))))........))))))..(((((((.....))))).))....))))))..)...---------....((((........)))). ( -29.86) >DroEre_CAF1 14169 120 - 1 GUAUCUUAUGUGAUGAAAUUUAUGCUUAAUUAAAAGCGACACAAGGCGUCACAAGCAACGCACAACGCAUUCGCCCAGAUAAGAACCGAAGGCACCAAAGGAACCAAGUUGGCCAUCGGC ...((((((((((((.......(((((......)))))........))))))..((((.((.....)).)).))....)))))).((((.(((......(....)......))).)))). ( -27.56) >DroYak_CAF1 14788 111 - 1 GUAUCUUAUGUGAUGAAAUUUAUGCUUAAUUAAAAGCGACACAAGGCGUCACAAGCAACGCACAACGCGUUCGCUCAGAUAAGACGCGAA---------AGAACCAAGCGGGCCAUCGGC (..((((((((((((.......(((((......)))))........)))))).((((((((.....))))).)))...))))))..)...---------....((....))(((...))) ( -28.36) >consensus GUAUCUUAUGUGAUGAAAUUUAUGCUUAAUUAAAAGCGACACAAGGCGUCACAAGCAACGCACAACGCGUUCGCCCAGAUAAGACGCGAA_________AGAACCAAGCGGGCCAUCGGC ((.((((((((((((.......(((((......)))))........))))))..(((((((.....))))).))....)))))).))........................(((...))) (-24.20 = -24.60 + 0.40)


| Location | 13,932,677 – 13,932,797 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -30.86 |
| Energy contribution | -30.62 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13932677 120 + 22224390 AUCUGGGCGAACGCGUUGUGCGUUGCUUGUGACGCCUUGUGUCGCUUUUAAUUAAGCAUAAAUUUCAUCACAUAAGAUACGAGAUGCAGUUAAAAGGGGCACGCAGUGGUGCCUUAUUUU ....(((((.((((.....((((((....))))))...(((((.(((((((((..((((...((..(((......)))..)).))))))))))))).))))))).))..)))))...... ( -35.60) >DroSec_CAF1 12632 120 + 1 AUCUGAGCGAACGCGUUGUGCGUUGCUUGUGACGCCUUGUGUCGCUUUUAAUUAAGCAUAAAUUUCAUCACAUAAGAUACGAGAUGCAGUUAAAAAGGGCACGCAGCGGUGCCUUAUUUU ....((((.(((((.....)))))))))(((((((...)))))))((((((((..((((...((..(((......)))..)).))))))))))))(((((((......)))))))..... ( -35.00) >DroSim_CAF1 16196 120 + 1 AUCUGGGCGAACGCGUUGUGCGUUGCUUGUGACGCCUUGUGUCGCUUUUAAUUAAGCAUAAAUUUCAUCACAUAAGAUACGAGAUGCAGUUAAAAAGGGCACGCAGCGGUGCCUUAUUUU ....(((((..(((....(((((.(((((((((((...)))))))((((((((..((((...((..(((......)))..)).)))))))))))).)))))))))))).)))))...... ( -34.20) >DroEre_CAF1 14209 119 + 1 AUCUGGGCGAAUGCGUUGUGCGUUGCUUGUGACGCCUUGUGUCGCUUUUAAUUAAGCAUAAAUUUCAUCACAUAAGAUACGAGCUGCAGUUAAGAGGGGCACGCAGCGCUGUUUGAGUU- .......(((((((((((((.(((.(((.((((((((((((((((((......))))....((........))..))))))))..)).)))).))).))).)))))))).)))))....- ( -37.60) >DroYak_CAF1 14819 120 + 1 AUCUGAGCGAACGCGUUGUGCGUUGCUUGUGACGCCUUGUGUCGCUUUUAAUUAAGCAUAAAUUUCAUCACAUAAGAUACGAGAUGCAGUUAAAAUGGGCACGCAGUGGUGCUUUAGUUU ....((((....(((..((((((.(((((((((((...)))))))((((((((..((((...((..(((......)))..)).)))))))))))).))))))))).)..)))....)))) ( -31.20) >consensus AUCUGGGCGAACGCGUUGUGCGUUGCUUGUGACGCCUUGUGUCGCUUUUAAUUAAGCAUAAAUUUCAUCACAUAAGAUACGAGAUGCAGUUAAAAAGGGCACGCAGCGGUGCCUUAUUUU ....((((.(((((.....)))))))))(((((((...)))))))((((((((..((((...((..(((......)))..)).))))))))))))(((((((......)))))))..... (-30.86 = -30.62 + -0.24)



| Location | 13,932,677 – 13,932,797 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -23.90 |
| Energy contribution | -24.90 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13932677 120 - 22224390 AAAAUAAGGCACCACUGCGUGCCCCUUUUAACUGCAUCUCGUAUCUUAUGUGAUGAAAUUUAUGCUUAAUUAAAAGCGACACAAGGCGUCACAAGCAACGCACAACGCGUUCGCCCAGAU .......(((((......)))))........(((..............(((((((.......(((((......)))))........))))))).(((((((.....))))).)).))).. ( -28.96) >DroSec_CAF1 12632 120 - 1 AAAAUAAGGCACCGCUGCGUGCCCUUUUUAACUGCAUCUCGUAUCUUAUGUGAUGAAAUUUAUGCUUAAUUAAAAGCGACACAAGGCGUCACAAGCAACGCACAACGCGUUCGCUCAGAU .......(((((......)))))........(((..............(((((((.......(((((......)))))........)))))))((((((((.....))))).)))))).. ( -29.76) >DroSim_CAF1 16196 120 - 1 AAAAUAAGGCACCGCUGCGUGCCCUUUUUAACUGCAUCUCGUAUCUUAUGUGAUGAAAUUUAUGCUUAAUUAAAAGCGACACAAGGCGUCACAAGCAACGCACAACGCGUUCGCCCAGAU .......(((...(((..((((((((........((((.((((...)))).)))).......(((((......)))))....)))).).))).)))(((((.....))))).)))..... ( -29.50) >DroEre_CAF1 14209 119 - 1 -AACUCAAACAGCGCUGCGUGCCCCUCUUAACUGCAGCUCGUAUCUUAUGUGAUGAAAUUUAUGCUUAAUUAAAAGCGACACAAGGCGUCACAAGCAACGCACAACGCAUUCGCCCAGAU -..........(((((((((..........)).)))))..........(((((((.......(((((......)))))........))))))).))...((.....))............ ( -24.16) >DroYak_CAF1 14819 120 - 1 AAACUAAAGCACCACUGCGUGCCCAUUUUAACUGCAUCUCGUAUCUUAUGUGAUGAAAUUUAUGCUUAAUUAAAAGCGACACAAGGCGUCACAAGCAACGCACAACGCGUUCGCUCAGAU ........((((......)))).........(((..............(((((((.......(((((......)))))........)))))))((((((((.....))))).)))))).. ( -26.46) >consensus AAAAUAAGGCACCGCUGCGUGCCCCUUUUAACUGCAUCUCGUAUCUUAUGUGAUGAAAUUUAUGCUUAAUUAAAAGCGACACAAGGCGUCACAAGCAACGCACAACGCGUUCGCCCAGAU .......(((((......)))))........(((..............(((((((.......(((((......)))))........))))))).(((((((.....))))).)).))).. (-23.90 = -24.90 + 1.00)

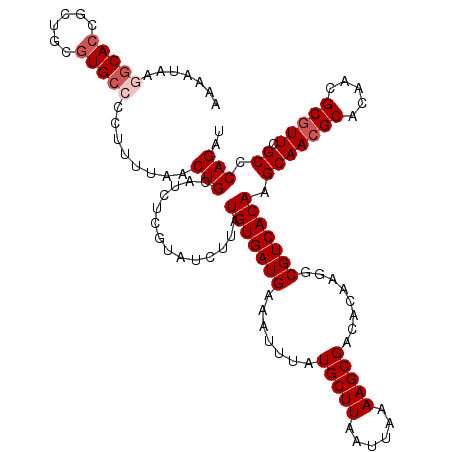

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:44 2006