
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,931,602 – 13,931,856 |
| Length | 254 |
| Max. P | 0.974476 |

| Location | 13,931,602 – 13,931,697 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.93 |
| Mean single sequence MFE | -29.34 |
| Consensus MFE | -26.34 |
| Energy contribution | -25.98 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13931602 95 + 22224390 AUAUUUGAAGCUAGUUGUGAACCUGAAUCUGGGGGCUGGGGCCAUGCAAUUAGUCUAAUUGCUAAGCAUGCCAGCCGGACUAUUUCCAUUUGCAU------------------------- ......((((.(((((.....(((......)))((((((.((...(((((((...)))))))...))...)))))).))))))))).........------------------------- ( -27.60) >DroSec_CAF1 11601 95 + 1 AUAUUUGAAGCUAGUUGCGAACCUGAAUCUGGGGGCUGGGGCCAUGCAAUUAGUCUAAUUGCUAAGCAUGCCAGCCGGACUAUUUCCAUUUGCAU------------------------- ...............(((((((((......)))((((((.((...(((((((...)))))))...))...))))))(((.....))).)))))).------------------------- ( -29.40) >DroSim_CAF1 15177 95 + 1 AUAUUUGAAGCUAGUUGCGAACCUGAAUCUGGGGGUUGGGGCCAUGCAAUUAGUAUAAUUGAUAAGCAUGCCAGCCGGAAUAUUUCCAUUUGCAU------------------------- ...............(((((((((......)))((((((...(((((.((((((...))))))..)))))))))))(((.....))).)))))).------------------------- ( -24.60) >DroEre_CAF1 13215 97 + 1 ACAUUUGAUGCUAGUUGUGAACCUGAAUCUGGGGGCUGGGGCCAUGCAAUUAGUCUAAUUGCUAAGCAUGCCAGCCGGACUAUUUCCAUUUGCAUU-----------------------C ......(((((....((.((((((......)))((((((.((...(((((((...)))))))...))...)))))).......)))))...)))))-----------------------. ( -28.60) >DroYak_CAF1 13750 120 + 1 AUAUUUGAAGCUAGUUGUGAACCUGAAUCUGGGGGCUGGGGCCAUGCAAUUAGUCUAAUUGCUAAGCAUGCCAGUCGGACUAUUUCCAUUUGCAUUCGGCCAAAAGCCCAGCCACCAUAC .........(((.........((.......))(((((..((((((((((.(((((..(((((.......).))))..))))).......))))))..))))...))))))))........ ( -36.50) >consensus AUAUUUGAAGCUAGUUGUGAACCUGAAUCUGGGGGCUGGGGCCAUGCAAUUAGUCUAAUUGCUAAGCAUGCCAGCCGGACUAUUUCCAUUUGCAU_________________________ ...............(((((((((......)))((((((.((...(((((((...)))))))...))...))))))(((.....))).)))))).......................... (-26.34 = -25.98 + -0.36)



| Location | 13,931,697 – 13,931,816 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.89 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -21.56 |
| Energy contribution | -22.16 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13931697 119 + 22224390 -CCAUCCCCCAUCCAUUCGCCAACCGAAUCUGGCCAAGACAGAUCGAAGAGGCAGCAGGCACAAACUGGCGCUAAUCGUUCAAACGAAAUCAGCAACAAUUCAUUUGCACAACAAGCGCC -.................(((...(((.((((.......)))))))....)))..............((((((..((((....)))).....((((........))))......)))))) ( -26.90) >DroSec_CAF1 11696 114 + 1 ----UCCCCCAUCCAUUCGCCAGCCGAAUCUGGCCAAGACAGAUCGAAGAGACAGCAGGC--AAACUGGCGCUAAUCGUUCAAACGAAAUCAGCAACAAUUCAUUUGCACAACAAGCGCC ----..............(((.(((((.((((.......)))))))........)).)))--.....((((((..((((....)))).....((((........))))......)))))) ( -27.20) >DroSim_CAF1 15272 114 + 1 ----UCCCCCGUCCAUUCGCCAGCCGAAUCUGGCCAAGACAGAUCGAAGAGGCAGCAGGC--AAACCGGCGCUAAUCGUUCAAACGAAAUCAGCAACAAUUCAUUUGCACAACAAGCGCC ----......(((..((((((((......)))))...((....)))))..)))....((.--...))((((((..((((....)))).....((((........))))......)))))) ( -28.60) >DroEre_CAF1 13312 117 + 1 CCCAUUCCCCAUCCAUUCCCCAACCGAAUCUGGCCAAGACAGAUCGAAGAGGCAGCAGGC--A-ACUGGCGCUAAUCGUUCAAACGAAAUCAGCAACAAUUCAUUUGCACAACAAGCGCC ..................((....(((.((((.......)))))))....))...(((..--.-.)))(((((..((((....)))).....((((........))))......))))). ( -25.10) >DroYak_CAF1 13870 118 + 1 CCCAUACCCCUUUCAUUCCCCAUCCAAAUCUGGCCAAGACAGAUCGAAGAGGCAGCAGGC--AAACUGGCGCUAAUCGUUCGAACGAAAUCAGCAACAAUUCAUUUGCACAACAAGCGCC ........(((......((.(.((...(((((.......))))).)).).))....))).--.....((((((..((((....)))).....((((........))))......)))))) ( -22.70) >consensus _CCAUCCCCCAUCCAUUCGCCAACCGAAUCUGGCCAAGACAGAUCGAAGAGGCAGCAGGC__AAACUGGCGCUAAUCGUUCAAACGAAAUCAGCAACAAUUCAUUUGCACAACAAGCGCC ..................(((...(((.((((.......)))))))..(......).))).......((((((..((((....)))).....((((........))))......)))))) (-21.56 = -22.16 + 0.60)



| Location | 13,931,697 – 13,931,816 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.89 |
| Mean single sequence MFE | -41.66 |
| Consensus MFE | -35.06 |
| Energy contribution | -35.58 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13931697 119 - 22224390 GGCGCUUGUUGUGCAAAUGAAUUGUUGCUGAUUUCGUUUGAACGAUUAGCGCCAGUUUGUGCCUGCUGCCUCUUCGAUCUGUCUUGGCCAGAUUCGGUUGGCGAAUGGAUGGGGGAUGG- ((((((.(((((.((((((((...........)))))))).))))).))))))(((........))).(((((((.((((((((..(((......)))..).).)))))).))))).))- ( -42.30) >DroSec_CAF1 11696 114 - 1 GGCGCUUGUUGUGCAAAUGAAUUGUUGCUGAUUUCGUUUGAACGAUUAGCGCCAGUUU--GCCUGCUGUCUCUUCGAUCUGUCUUGGCCAGAUUCGGCUGGCGAAUGGAUGGGGGA---- ((((((.(((((.((((((((...........)))))))).))))).)))))).....--..((.((((((.(((((....))...(((((......)))))))).)))))).)).---- ( -42.90) >DroSim_CAF1 15272 114 - 1 GGCGCUUGUUGUGCAAAUGAAUUGUUGCUGAUUUCGUUUGAACGAUUAGCGCCGGUUU--GCCUGCUGCCUCUUCGAUCUGUCUUGGCCAGAUUCGGCUGGCGAAUGGACGGGGGA---- ((((((.(((((.((((((((...........)))))))).))))).))))))((...--.)).....((((.((.((.((((..((((......)))))))).)).)).))))..---- ( -42.80) >DroEre_CAF1 13312 117 - 1 GGCGCUUGUUGUGCAAAUGAAUUGUUGCUGAUUUCGUUUGAACGAUUAGCGCCAGU-U--GCCUGCUGCCUCUUCGAUCUGUCUUGGCCAGAUUCGGUUGGGGAAUGGAUGGGGAAUGGG ((((((.(((((.((((((((...........)))))))).))))).))))))...-.--.((((...((((.((.((...(((..(((......)))..))).)).)).))))..)))) ( -43.40) >DroYak_CAF1 13870 118 - 1 GGCGCUUGUUGUGCAAAUGAAUUGUUGCUGAUUUCGUUCGAACGAUUAGCGCCAGUUU--GCCUGCUGCCUCUUCGAUCUGUCUUGGCCAGAUUUGGAUGGGGAAUGAAAGGGGUAUGGG ((((((.(((((.(.((((((...........)))))).).))))).)))))).....--.((((.((((((((((.(((((((...........)))))))...)).)))))))))))) ( -36.90) >consensus GGCGCUUGUUGUGCAAAUGAAUUGUUGCUGAUUUCGUUUGAACGAUUAGCGCCAGUUU__GCCUGCUGCCUCUUCGAUCUGUCUUGGCCAGAUUCGGCUGGCGAAUGGAUGGGGGAUGG_ ((((((.(((((.((((((((...........)))))))).))))).))))))(((........))).((((.((.((.((((..((((......)))))))).)).)).))))...... (-35.06 = -35.58 + 0.52)



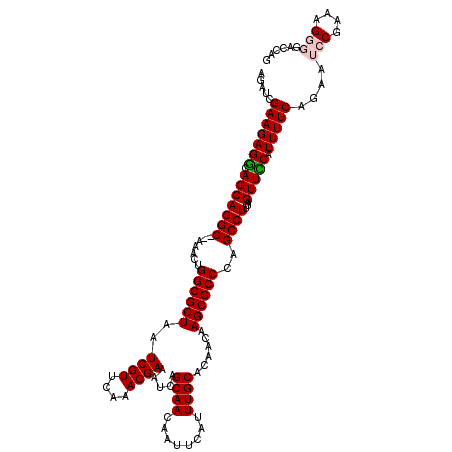
| Location | 13,931,736 – 13,931,856 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.20 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -27.58 |
| Energy contribution | -27.98 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13931736 120 + 22224390 AGAUCGAAGAGGCAGCAGGCACAAACUGGCGCUAAUCGUUCAAACGAAAUCAGCAACAAUUCAUUUGCACAACAAGCGCCCAGCCUUUAGUUCCAUUUUCAGAAUCCGAAAGGGGUCCAG .(((((((((((.(((((((.......((((((..((((....)))).....((((........))))......))))))..))))...))))).))))).....((....))))))... ( -34.10) >DroSec_CAF1 11732 118 + 1 AGAUCGAAGAGACAGCAGGC--AAACUGGCGCUAAUCGUUCAAACGAAAUCAGCAACAAUUCAUUUGCACAACAAGCGCCCAGCCUUUAGUUUCAUUUUCAGAAUCCGAAAGGGGACCAG .(.((...(((((...((((--.....((((((..((((....)))).....((((........))))......))))))..))))...)))))..........(((....))))).).. ( -31.80) >DroSim_CAF1 15308 118 + 1 AGAUCGAAGAGGCAGCAGGC--AAACCGGCGCUAAUCGUUCAAACGAAAUCAGCAACAAUUCAUUUGCACAACAAGCGCCCAGCCUUUAGUUUCAUUUUCAGAAUCCGAAAGGGGACCAG .((....((((((....((.--...))((((((..((((....)))).....((((........))))......))))))..))))))....))..........(((....)))...... ( -31.80) >DroEre_CAF1 13352 117 + 1 AGAUCGAAGAGGCAGCAGGC--A-ACUGGCGCUAAUCGUUCAAACGAAAUCAGCAACAAUUCAUUUGCACAACAAGCGCCCAGCCUGUGGUUCCAUUUUCAGAAACCGAAAGAGGCCCAG ........(.(((.((((((--.-...((((((..((((....)))).....((((........))))......))))))..))))))((((.(.......).)))).......)))).. ( -34.40) >DroYak_CAF1 13910 118 + 1 AGAUCGAAGAGGCAGCAGGC--AAACUGGCGCUAAUCGUUCGAACGAAAUCAGCAACAAUUCAUUUGCACAACAAGCGCCCAGCCUUUAGUUCCAUUUUCAGAAACCGAAAGAGGACCAG .......((((((...((..--...))((((((..((((....)))).....((((........))))......))))))..)))))).(.(((.(((((.......))))).))).).. ( -30.20) >consensus AGAUCGAAGAGGCAGCAGGC__AAACUGGCGCUAAUCGUUCAAACGAAAUCAGCAACAAUUCAUUUGCACAACAAGCGCCCAGCCUUUAGUUCCAUUUUCAGAAUCCGAAAGGGGACCAG .....(((((((.(((((((.......((((((..((((....)))).....((((........))))......))))))..))))...))))).)))))....(((....)))...... (-27.58 = -27.98 + 0.40)


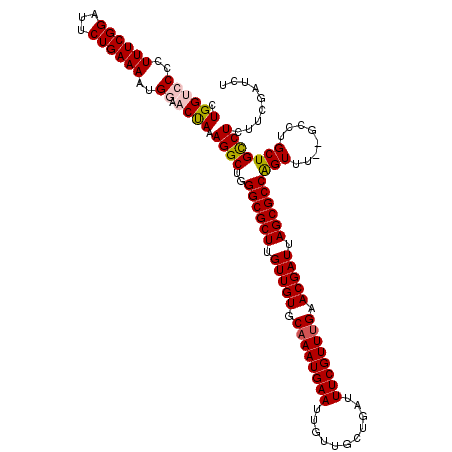
| Location | 13,931,736 – 13,931,856 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.20 |
| Mean single sequence MFE | -37.90 |
| Consensus MFE | -32.42 |
| Energy contribution | -32.94 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13931736 120 - 22224390 CUGGACCCCUUUCGGAUUCUGAAAAUGGAACUAAAGGCUGGGCGCUUGUUGUGCAAAUGAAUUGUUGCUGAUUUCGUUUGAACGAUUAGCGCCAGUUUGUGCCUGCUGCCUCUUCGAUCU ..((.((..((((((...))))))..))......((((..((((((.(((((.((((((((...........)))))))).))))).)))))).......))))....)).......... ( -35.00) >DroSec_CAF1 11732 118 - 1 CUGGUCCCCUUUCGGAUUCUGAAAAUGAAACUAAAGGCUGGGCGCUUGUUGUGCAAAUGAAUUGUUGCUGAUUUCGUUUGAACGAUUAGCGCCAGUUU--GCCUGCUGUCUCUUCGAUCU ..((((...((((((...))))))..((.((...((((..((((((.(((((.((((((((...........)))))))).))))).)))))).....--))))...)).))...)))). ( -38.10) >DroSim_CAF1 15308 118 - 1 CUGGUCCCCUUUCGGAUUCUGAAAAUGAAACUAAAGGCUGGGCGCUUGUUGUGCAAAUGAAUUGUUGCUGAUUUCGUUUGAACGAUUAGCGCCGGUUU--GCCUGCUGCCUCUUCGAUCU ..((((...((((((...))))))..........((((..((((((.(((((.((((((((...........)))))))).))))).))))))((...--.))....))))....)))). ( -36.40) >DroEre_CAF1 13352 117 - 1 CUGGGCCUCUUUCGGUUUCUGAAAAUGGAACCACAGGCUGGGCGCUUGUUGUGCAAAUGAAUUGUUGCUGAUUUCGUUUGAACGAUUAGCGCCAGU-U--GCCUGCUGCCUCUUCGAUCU ..((((.......((((((.......)))))).(((((..((((((.(((((.((((((((...........)))))))).))))).))))))...-.--)))))..))))......... ( -45.30) >DroYak_CAF1 13910 118 - 1 CUGGUCCUCUUUCGGUUUCUGAAAAUGGAACUAAAGGCUGGGCGCUUGUUGUGCAAAUGAAUUGUUGCUGAUUUCGUUCGAACGAUUAGCGCCAGUUU--GCCUGCUGCCUCUUCGAUCU .((((((..((((((...))))))..)).)))).((((.((((((.......))....((((((.((((((..((((....)))))))))).))))))--))))...))))......... ( -34.70) >consensus CUGGUCCCCUUUCGGAUUCUGAAAAUGGAACUAAAGGCUGGGCGCUUGUUGUGCAAAUGAAUUGUUGCUGAUUUCGUUUGAACGAUUAGCGCCAGUUU__GCCUGCUGCCUCUUCGAUCU .((((((..((((((...))))))..)).)))).((((..((((((.(((((.((((((((...........)))))))).))))).))))))(((........)))))))......... (-32.42 = -32.94 + 0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:40 2006