
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,930,855 – 13,931,249 |
| Length | 394 |
| Max. P | 0.984441 |

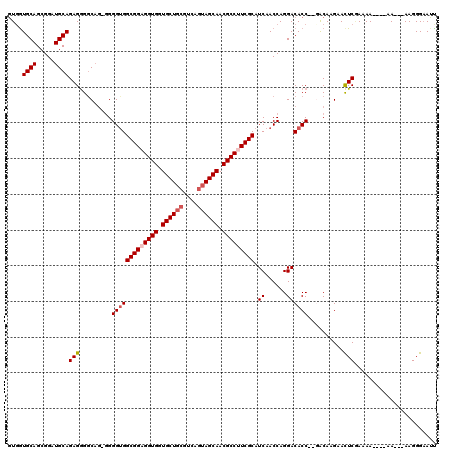
| Location | 13,930,855 – 13,930,967 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.62 |
| Mean single sequence MFE | -38.74 |
| Consensus MFE | -31.55 |
| Energy contribution | -32.39 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13930855 112 - 22224390 GUUGUGCAGCGGAUGCAGAGAGGCAG-AGGGUGGCGGAGGUGGUGCUGCGUCAGUAGCAACGCCUUCGCAUCAACCAGGACACCAGGACAAGAAUUCGAAAA----AA---AAGGGAAUU .((((.(..(...(((......))).-.)(((((((((((((.((((((....)))))).))))))))).((......)))))).).)))).(((((.....----..---....))))) ( -37.80) >DroSec_CAF1 10903 111 - 1 GUGGUGCAGCGGAUGCAGAGGGGCAG-GGGGUGGCGGAGGUGGUGCUGCGUCAAUAGCAACGCCUUCGCAUCAACCAGGACACC--GACAAGAACUCGAAAAA---AA---AAGGGAAUU (((((((......(((......)))(-(..(..(((((((((.(((((......))))).)))))))))..)..))..).))))--.)).....(((......---..---..))).... ( -35.50) >DroSim_CAF1 13501 110 - 1 GUGGUGCAGCGGAUGCAGAGGGGCAG-GGGGUGGCGGAGGUGGUGCUGCGUCAGUAGCAACGCCUUCGCAUCAACCAGGACACC--GACAAGAACUCGAAAA----AA---AAGGGAAUU (((((((......(((......)))(-(..(..(((((((((.((((((....)))))).)))))))))..)..))..).))))--.)).....(((.....----..---..))).... ( -40.00) >DroEre_CAF1 12636 112 - 1 UUGGUGCAGCGGUUGCAGAGGGGC--GGGGGCGGCGGUGGUGUUGCUGCGUCAGGAGCAACGCCUUCGCAUCAACCAGGACACC--CGCAAGAACUCCAAAA----AAAGGAAGGGCAAC ...........(((((......((--(((....((((.(((((((((.(....).))))))))).)))).((......))..))--)))......(((....----...)))...))))) ( -40.80) >DroYak_CAF1 13001 118 - 1 GUGAUGCAGCGGAUGCAGAGGGUCAGGGGGGUGGCGGCGGUGGUGCUGCGUCAGUAGCAACGCCUUCGCAUCAACCAGGACACC--CACAAGAACUCAAAAAAAAAAAAAGAAGGGAAUU (((.((((.....))))..(((((..((..(..((((.((((.((((((....)))))).)))).))))..)..))..))).))--)))............................... ( -39.60) >consensus GUGGUGCAGCGGAUGCAGAGGGGCAG_GGGGUGGCGGAGGUGGUGCUGCGUCAGUAGCAACGCCUUCGCAUCAACCAGGACACC__GACAAGAACUCGAAAA____AA___AAGGGAAUU ....((((.....))))(((.........(((((((((((((.((((((....)))))).))))))))).((......))))))..........)))....................... (-31.55 = -32.39 + 0.84)


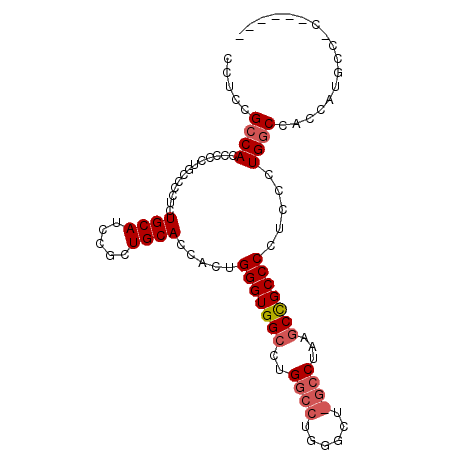
| Location | 13,930,928 – 13,931,027 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 79.38 |
| Mean single sequence MFE | -27.96 |
| Consensus MFE | -18.87 |
| Energy contribution | -20.40 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13930928 99 + 22224390 CCUCCGCCACCCUCUGCCUCUCUGCAUCCGCUGCACAACUGGGUGGCCUGGCCUGGGCUCGCCUAAACUGCCCCUCGCUGACCACCCUGCCACGUUGCC .......................(((..((..(((.....((((((.(.(((..((((...........))))...)))).)))))))))..)).))). ( -26.00) >DroSec_CAF1 10975 83 + 1 CCUCCGCCACCCCCUGCCCCUCUGCAUCCGCUGCACCACUGGGUGGCCUG----------GCCUAAGCCGCCCCUCCCUGGCCACCAUGCCCC------ .....(((((((..((......((((.....)))).))..))))))).((----------(((................))))).........------ ( -23.09) >DroSim_CAF1 13572 88 + 1 CCUCCGCCACCCCCUGCCCCUCUGCAUCCGCUGCACCACUGGGUGGCCUGGCCUGGGCUGGCCUAAGCCGCCCCUCCCUGGCCACCAU----------- .....(((((((..((......((((.....)))).))..))))))).(((((.((((.(((....)))))))......)))))....----------- ( -34.80) >consensus CCUCCGCCACCCCCUGCCCCUCUGCAUCCGCUGCACCACUGGGUGGCCUGGCCUGGGCU_GCCUAAGCCGCCCCUCCCUGGCCACCAUGCC_C______ .....((((.............((((.....)))).....(((((((..((((......))))...))))))).....))))................. (-18.87 = -20.40 + 1.53)

| Location | 13,930,928 – 13,931,027 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 79.38 |
| Mean single sequence MFE | -37.53 |
| Consensus MFE | -24.61 |
| Energy contribution | -24.83 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13930928 99 - 22224390 GGCAACGUGGCAGGGUGGUCAGCGAGGGGCAGUUUAGGCGAGCCCAGGCCAGGCCACCCAGUUGUGCAGCGGAUGCAGAGAGGCAGAGGGUGGCGGAGG .....(((.((.((((((((.((...((((.((....))..))))..))..))))))))..((.((((.....)))).)).........)).))).... ( -38.00) >DroSec_CAF1 10975 83 - 1 ------GGGGCAUGGUGGCCAGGGAGGGGCGGCUUAGGC----------CAGGCCACCCAGUGGUGCAGCGGAUGCAGAGGGGCAGGGGGUGGCGGAGG ------..(((..(((.(((.......))).)))...))----------)..(((((((..((.(.(.((....))...).).))..)))))))..... ( -34.30) >DroSim_CAF1 13572 88 - 1 -----------AUGGUGGCCAGGGAGGGGCGGCUUAGGCCAGCCCAGGCCAGGCCACCCAGUGGUGCAGCGGAUGCAGAGGGGCAGGGGGUGGCGGAGG -----------....(((((.(((..((.(......).))..))).))))).(((((((..((.(.(.((....))...).).))..)))))))..... ( -40.30) >consensus ______G_GGCAUGGUGGCCAGGGAGGGGCGGCUUAGGC_AGCCCAGGCCAGGCCACCCAGUGGUGCAGCGGAUGCAGAGGGGCAGGGGGUGGCGGAGG .............(((.(((.......))).)))..(((........)))..(((((((.((......))...(((......)))..)))))))..... (-24.61 = -24.83 + 0.23)


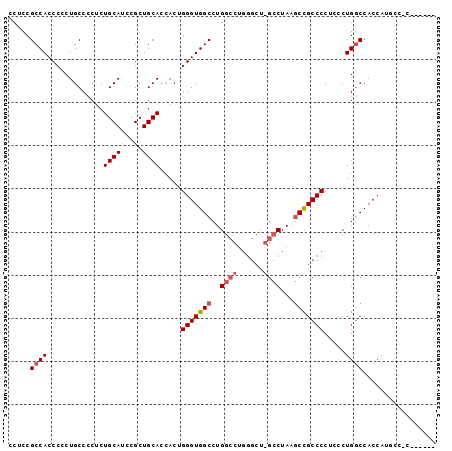
| Location | 13,930,967 – 13,931,067 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 78.91 |
| Mean single sequence MFE | -38.83 |
| Consensus MFE | -25.85 |
| Energy contribution | -27.63 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13930967 100 + 22224390 UGGGUGGCCUGGCCUGGGCUCGCCUAAACUGCCCCUCGCUGACCACCCUGCCACGUUGCCACGAGAGUCAGCAACUCCACUGGCGACUCCAAGCCAUCCG .(((((((.(((...((((...........))))....................(((((((...((((.....))))...))))))).))).))))))). ( -36.90) >DroSec_CAF1 11014 84 + 1 UGGGUGGCCUG----------GCCUAAGCCGCCCCUCCCUGGCCACCAUGCCCC------GGGAGAGCCAGCAACUCCACUGGCGACUCCAACCCAUCCG ((((((((..(----------((....)))))).(((((.(((......)))..------))))).(((((........))))).......))))).... ( -34.00) >DroSim_CAF1 13611 89 + 1 UGGGUGGCCUGGCCUGGGCUGGCCUAAGCCGCCCCUCCCUGGCCACCAU-----------GGGAGAGCCAGCAACUCCACUGGCGACUCCAACCCAUCCG ..(((((((.((...((((.(((....)))))))...)).)))))))..-----------.((((.(((((........)))))..)))).......... ( -45.60) >consensus UGGGUGGCCUGGCCUGGGCU_GCCUAAGCCGCCCCUCCCUGGCCACCAUGCC_C______GGGAGAGCCAGCAACUCCACUGGCGACUCCAACCCAUCCG ..(((((((......((((...........))))......)))))))..............((((.(((((........)))))..)))).......... (-25.85 = -27.63 + 1.78)



| Location | 13,930,967 – 13,931,067 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 78.91 |
| Mean single sequence MFE | -45.30 |
| Consensus MFE | -28.38 |
| Energy contribution | -28.83 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13930967 100 - 22224390 CGGAUGGCUUGGAGUCGCCAGUGGAGUUGCUGACUCUCGUGGCAACGUGGCAGGGUGGUCAGCGAGGGGCAGUUUAGGCGAGCCCAGGCCAGGCCACCCA .((.((((((((..((((((.((...(((((((((..(.((.(.....).)).)..)))))))))....)).)...)))))..))))))))..))..... ( -45.10) >DroSec_CAF1 11014 84 - 1 CGGAUGGGUUGGAGUCGCCAGUGGAGUUGCUGGCUCUCCC------GGGGCAUGGUGGCCAGGGAGGGGCGGCUUAGGC----------CAGGCCACCCA .((.(((.((((((((((((((......)))).(((((((------..(((......))).)))))))))))))....)----------))).))).)). ( -42.70) >DroSim_CAF1 13611 89 - 1 CGGAUGGGUUGGAGUCGCCAGUGGAGUUGCUGGCUCUCCC-----------AUGGUGGCCAGGGAGGGGCGGCUUAGGCCAGCCCAGGCCAGGCCACCCA ...(((((..(((...((((((......))))))))))))-----------))(((((((..((..(((((((....))).))))...)).))))))).. ( -48.10) >consensus CGGAUGGGUUGGAGUCGCCAGUGGAGUUGCUGGCUCUCCC______G_GGCAUGGUGGCCAGGGAGGGGCGGCUUAGGC_AGCCCAGGCCAGGCCACCCA .........(((.((.(((.........((((.(((((((........(((......))).))))))).))))...(((........))).))).))))) (-28.38 = -28.83 + 0.45)

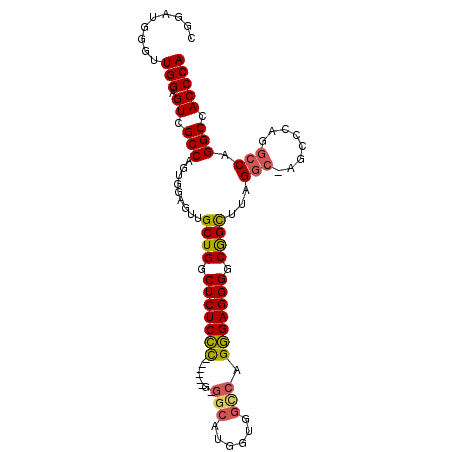

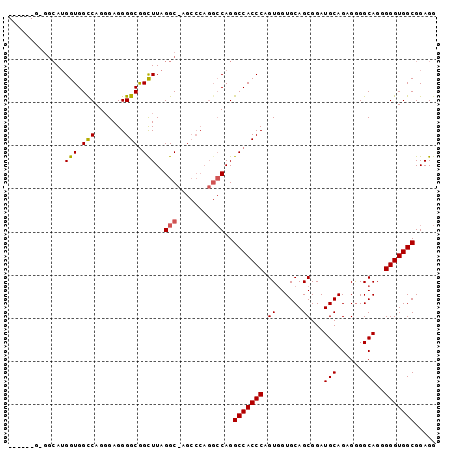
| Location | 13,931,027 – 13,931,147 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.11 |
| Mean single sequence MFE | -45.42 |
| Consensus MFE | -34.09 |
| Energy contribution | -34.77 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13931027 120 + 22224390 ACGAGAGUCAGCAACUCCACUGGCGACUCCAAGCCAUCCGGAUUCAGCCCAUACGACGCAUGCACAGAGCCAUCGUUGGAGUGGCAUGUGUGGCAAGUGUCAUGGGUGGUGAUCUCAGCA ..((((.(((.(...(((..((((........))))...)))....((((((..(((((.((((((.((((((.......))))).).))).))).)))))))))))).))))))).... ( -45.30) >DroSec_CAF1 11058 120 + 1 GGGAGAGCCAGCAACUCCACUGGCGACUCCAACCCAUCCGGCUUCAGCCCAUACGACGCAUGCACAGAGCCAUCGUUUGAGUGGCAUGUGUGGCAAAUGUCAUGGGUGGGGAUCUCUGCA .((((.(((((........)))))..))))..((((((((((....))).....((((..((((((.((((((.......))))).).))).)))..))))..))))))).......... ( -49.20) >DroSim_CAF1 13660 120 + 1 GGGAGAGCCAGCAACUCCACUGGCGACUCCAACCCAUCCGGCUUCAGCCCAUACGACGCAUGCACAGAGCCAUCGUUGGAGUGGCAUGUGUGGCAAGUGUCAUGGGUGGGGAUCUCUGCA .((((.(((((........)))))..))))..((((((((((....))).....(((((.((((((.((((((.......))))).).))).))).)))))..))))))).......... ( -53.10) >DroYak_CAF1 13175 108 + 1 GGGAAGGCGAGCAACUCCAGUGGCGACUCCAACCCAUACGGAUUCAGCCCAAACGACGCAUGCACAGAGCCAUCGCUGGAGUGGCAUGUGUUCCG------------GGGGAUUUUAGCA .((((.(((.((.(((((((((((((.(((.........))).)).((.........)).........)))...)))))))).)).))).)))).------------............. ( -34.10) >consensus GGGAGAGCCAGCAACUCCACUGGCGACUCCAACCCAUCCGGAUUCAGCCCAUACGACGCAUGCACAGAGCCAUCGUUGGAGUGGCAUGUGUGGCAAGUGUCAUGGGUGGGGAUCUCAGCA ..(((((((((........)))))........(((((((((......))....(.((((((((.((...(((....)))..)))))))))).)..........)))))))..)))).... (-34.09 = -34.77 + 0.69)

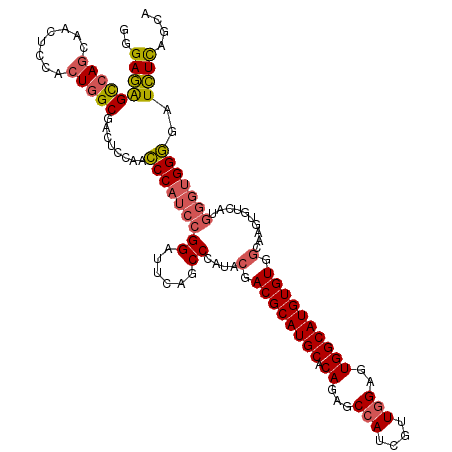

| Location | 13,931,147 – 13,931,249 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -29.86 |
| Consensus MFE | -21.94 |
| Energy contribution | -22.08 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.939077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13931147 102 - 22224390 -GUGCGCCCUGUGCACCCUGCAGCUUGCGCCUAUUGCAAUAUAUGCCUGCAGAUUGGUCAGUUCAAACACUGACAUCCGGCGGAACUCCCCCUC-----------------UCCAUCCCC -((((((...)))))).((((((((((((.....))))).....).))))))....((((((......))))))....((.((....)).))..-----------------......... ( -28.90) >DroSec_CAF1 11178 101 - 1 -GUGCGCCCUGUGCACCCUGCAGCUUGCGCCUAUUGCAAUAUAUGCCUGCAGAUUGGUCAGUUCAAACACUGACAUCCGGCGGAACC--CCCUGCA---------------CCCA-CCCC -((((((...))))))..(((((.((.((((..(((((.........)))))....((((((......))))))....)))).))..--..)))))---------------....-.... ( -30.70) >DroSim_CAF1 13780 100 - 1 NNNNNNNNNNNNNNNNCCUGCAGCUUGCGCCUAUUGCAAUAUAUGCCUGCAGAUUGGUCAGUUCAAACACUGACAUCCGGCGGAACC--CCCUC-----------------UCCA-GCCC .................((((((((((((.....))))).....).))))))....((((((......))))))....((((((...--.....-----------------))).-))). ( -25.10) >DroEre_CAF1 12852 111 - 1 -GUGCGCCCUGUGCACCCUGCAAGUUGCGCCUAUUGCAAUAUAUGCCUGCAGAUUGGUCAGUUCAAACACUGACAUCCGGUGGCAGC--CCCUCCACCCC-----GCACUUGCCA-CACA -((((((...))))))...((((((.(((....(((((.........)))))....((((((......))))))....(((((.((.--..))))))).)-----))))))))..-.... ( -35.30) >DroYak_CAF1 13283 116 - 1 -GUGCGCCCUGUGCACCCUGCAGCUUGCGCCUAUUGCAAUAUAUGCCUGCAGAUUGGUCAGUUCAAACACUGACAUCCGGCGGAAUC--CCCUCUCCCCCCCCUCCCCCUCUCCA-CCCC -((((((...)))))).((((((((((((.....))))).....).))))))....((((((......))))))....((.(((...--.....)))))................-.... ( -29.30) >consensus _GUGCGCCCUGUGCACCCUGCAGCUUGCGCCUAUUGCAAUAUAUGCCUGCAGAUUGGUCAGUUCAAACACUGACAUCCGGCGGAACC__CCCUC_________________UCCA_CCCC .((((((...))))))((.((.....))(((..(((((.........)))))....((((((......))))))....)))))..................................... (-21.94 = -22.08 + 0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:36 2006