
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,929,966 – 13,930,096 |
| Length | 130 |
| Max. P | 0.563588 |

| Location | 13,929,966 – 13,930,060 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.09 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -15.31 |
| Energy contribution | -15.91 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
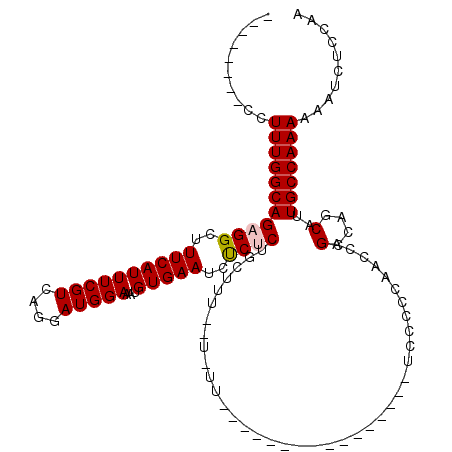
>X_DroMel_CAF1 13929966 94 + 22224390 CCCCAAAGCAUAGCACC------AAAAAAAAGGGACCCCC--------CCUUUGGCAGAGGCUUUCAUUUCGUCAGGAUGGAAAUGUGAAUCUCUCUGCUUUCUUUUU------------ .................------.....((((((.....)--------)))))((((((((..((((((((((....)))))...)))))..))))))))........------------ ( -24.80) >DroSec_CAF1 10008 90 + 1 CCCCAAAGCAUCGCAGC------AAAAA-AAGGGACC-CC--------CCUUUGGCAGAGGCUUUCAUUUCGUCAGGAUGGAAAUGUGAAUCCCUCUGCUUU--UUUU------------ .......((......))------....(-(((((...-.)--------)))))((((((((..((((((((((....)))))...)))))..))))))))..--....------------ ( -28.60) >DroSim_CAF1 12588 89 + 1 CCCCAAAGCAUCGCAGC------AAAAA-AAGGGACC-CC--------CCUUUGGCAGAGGCUUUCAUUUCGUCAGGAUGGAAAUGUGAAUCCCUCUGCUUU--U-UU------------ .......((......))------....(-(((((...-.)--------)))))((((((((..((((((((((....)))))...)))))..))))))))..--.-..------------ ( -28.60) >DroEre_CAF1 11734 98 + 1 CCCCAAAGCAGCGCAGC------AAAAAAAUGGGAGC-CCUCCCCACUCCUUUGGCAGAGGCUUUCAUUUCGUCAGGAUGGAAAUGUGAAUCUCCCGCCUUU--U-UC------------ .......((......))------....(((((..(((-(.((.(((......)))..))))))..)))))....(((..(((.((....)).)))..)))..--.-..------------ ( -22.50) >DroYak_CAF1 12064 108 + 1 CCCCAAAGCAUCGCAGCGCAAAAAAAAAAAAGGGACC-CC--------CCUUUGGCAGAGGCUUUCAUUUCGUCAGAAUGGAAAUGUGAAUCUCCCUUUUUU--U-UUUCCUUUAACCCC .......((......))....((((((((((((((..-..--------((((.....)))).((((((((.....)))))))).........))))))))))--)-)))........... ( -22.50) >consensus CCCCAAAGCAUCGCAGC______AAAAAAAAGGGACC_CC________CCUUUGGCAGAGGCUUUCAUUUCGUCAGGAUGGAAAUGUGAAUCUCUCUGCUUU__U_UU____________ .(((...........................)))...................((((((((..((((((((((....)))))...)))))..)))))))).................... (-15.31 = -15.91 + 0.60)


| Location | 13,930,000 – 13,930,096 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.29 |
| Mean single sequence MFE | -20.40 |
| Consensus MFE | -16.40 |
| Energy contribution | -16.56 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13930000 96 + 22224390 --------CCUUUGGCAGAGGCUUUCAUUUCGUCAGGAUGGAAAUGUGAAUCUCUCUGCUUUCUUUUU----------------UCCCCCAACCAGCCAGCAUUGCCAAAAAAUUUCCAA --------..(((((((((((..((((((((((....)))))...)))))..))))((((..((....----------------..........))..)))).))))))).......... ( -20.04) >DroSec_CAF1 10040 94 + 1 --------CCUUUGGCAGAGGCUUUCAUUUCGUCAGGAUGGAAAUGUGAAUCCCUCUGCUUU--UUUU----------------UCCCCCAACCAGCCAGCAUUGCCAAAAAAUCUCCAA --------..(((((((((((..((((((((((....)))))...)))))..))))((((..--....----------------..............)))).))))))).......... ( -21.85) >DroSim_CAF1 12620 93 + 1 --------CCUUUGGCAGAGGCUUUCAUUUCGUCAGGAUGGAAAUGUGAAUCCCUCUGCUUU--U-UU----------------UCCCCCAACCAGCCAGCAUUGCCAAAAAAUCUCCAA --------..(((((((((((..((((((((((....)))))...)))))..))))((((..--.-..----------------..............)))).))))))).......... ( -21.91) >DroEre_CAF1 11767 103 + 1 UCCCCACUCCUUUGGCAGAGGCUUUCAUUUCGUCAGGAUGGAAAUGUGAAUCUCCCGCCUUU--U-UC--------------CCACCCCCAACCAGCCAGCAUUGCCAAAAAAUCUCCAG ..........((((((((.((((.....(((....))).(((((.(((.......)))...)--)-))--------------)...........))))....)))))))).......... ( -18.60) >DroYak_CAF1 12103 109 + 1 --------CCUUUGGCAGAGGCUUUCAUUUCGUCAGAAUGGAAAUGUGAAUCUCCCUUUUUU--U-UUUCCUUUAACCCCCACCACCCCAAGCCAGCGAGCAUUGCCAAAAAAUCUCCAA --------..((((((((.(((((....(((....))).(((((...(((........))).--.-)))))..................))))).(....).)))))))).......... ( -19.60) >consensus ________CCUUUGGCAGAGGCUUUCAUUUCGUCAGGAUGGAAAUGUGAAUCUCUCUGCUUU__U_UU________________UCCCCCAACCAGCCAGCAUUGCCAAAAAAUCUCCAA ..........(((((((((((..((((((((((....)))))...)))))..)))).......................................(....)..))))))).......... (-16.40 = -16.56 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:29 2006