
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,928,952 – 13,929,073 |
| Length | 121 |
| Max. P | 0.999924 |

| Location | 13,928,952 – 13,929,046 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.08 |
| Mean single sequence MFE | -43.43 |
| Consensus MFE | -25.92 |
| Energy contribution | -26.86 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.60 |
| SVM decision value | 4.18 |
| SVM RNA-class probability | 0.999826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
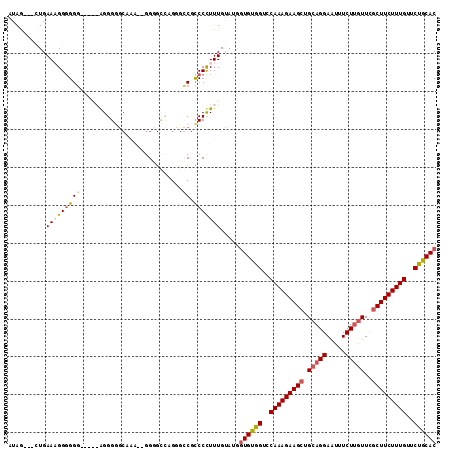
>X_DroMel_CAF1 13928952 94 + 22224390 AUAG---GUGAA-GGGGGG-----AGGGAG------GGG----GUUCCGUCCUUUUGUAUGGUGUGGUCCAAAGAAGCUGCAGGAGUUUCUUAUUUUCUUCUUUGUUCUGCAC ....---.....-((((((-----(.((((------...----.)))).))))))).....(((..(..((((((((....(((.....))).....))))))))..)..))) ( -28.20) >DroSec_CAF1 9038 103 + 1 AUAG---GUGAAAGGGGGG-----AGGGGGCAAA--GGGGCCAGGGCCGCCCUUUUGUAUGGUGUGGUCCAAAGAAGCUGCAGGAAUUUCUUGUUCGCUUCUUUGUUCUGCAC ....---(..((((((.((-----....(((...--...)))....)).))))))..)...(((..(..(((((((((.(((((.....)))))..)))))))))..)..))) ( -46.70) >DroEre_CAF1 10783 113 + 1 AUAGGUGCUGAAAGGGGGCGCCAAAGGGUGCGGGGGGGUGUCGGUGGCGCCCCUUUGUACGGUGCGGUCCAAAGAAGCUGCAGGAAUUUCUGGUUCGCUUCUUUGUUCCGCAC ...((((((.......)))))).....((((((((((((((.....)))))))))))))).((((((..(((((((((.((.((.....)).))..)))))))))..)))))) ( -59.70) >DroYak_CAF1 11024 103 + 1 AUAA---CUGAAUGGCGGGGGGGAGGGGGUCAAA--GGAGCAAGGGGCGCCCCUUUG-----UGUGGUCCAAAGAAGCUGCAGGAAUUUCUUGUUCGCUUCUUUGUUCUGCAC ....---(((.....)))...((((((((((...--.........))).)))))))(-----((..(..(((((((((.(((((.....)))))..)))))))))..)..))) ( -39.10) >consensus AUAG___CUGAAAGGGGGG_____AGGGGGCAAA__GGGGCCAGGGCCGCCCCUUUGUAUGGUGUGGUCCAAAGAAGCUGCAGGAAUUUCUUGUUCGCUUCUUUGUUCUGCAC ..........(((((((((...........................)).))))))).....((((((..(((((((((.(((((.....)))))..)))))))))..)))))) (-25.92 = -26.86 + 0.94)


| Location | 13,928,952 – 13,929,046 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.08 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -15.12 |
| Energy contribution | -15.00 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
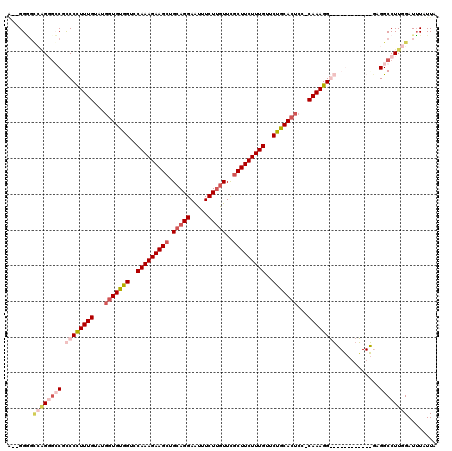
>X_DroMel_CAF1 13928952 94 - 22224390 GUGCAGAACAAAGAAGAAAAUAAGAAACUCCUGCAGCUUCUUUGGACCACACCAUACAAAAGGACGGAAC----CCC------CUCCCU-----CCCCCC-UUCAC---CUAU (((.((..((((((((......((......))....)))))))).................(((.(((..----...------.))).)-----))...)-).)))---.... ( -18.70) >DroSec_CAF1 9038 103 - 1 GUGCAGAACAAAGAAGCGAACAAGAAAUUCCUGCAGCUUCUUUGGACCACACCAUACAAAAGGGCGGCCCUGGCCCC--UUUGCCCCCU-----CCCCCCUUUCAC---CUAU (((..(..(((((((((.....((......))...)))))))))..)..)))......((((((.((....(((...--...)))....-----)).))))))...---.... ( -28.80) >DroEre_CAF1 10783 113 - 1 GUGCGGAACAAAGAAGCGAACCAGAAAUUCCUGCAGCUUCUUUGGACCGCACCGUACAAAGGGGCGCCACCGACACCCCCCCGCACCCUUUGGCGCCCCCUUUCAGCACCUAU ((((((..(((((((((....(((......)))..)))))))))..)))))).((..((((((((((((.....................))))).)))))))..))...... ( -46.50) >DroYak_CAF1 11024 103 - 1 GUGCAGAACAAAGAAGCGAACAAGAAAUUCCUGCAGCUUCUUUGGACCACA-----CAAAGGGGCGCCCCUUGCUCC--UUUGACCCCCUCCCCCCCGCCAUUCAG---UUAU (((.....(((((((((.....((......))...)))))))))...))).-----((((((((((.....))))))--)))).......................---.... ( -28.90) >consensus GUGCAGAACAAAGAAGCGAACAAGAAAUUCCUGCAGCUUCUUUGGACCACACCAUACAAAAGGGCGCCACUGGCCCC__UUUGCCCCCU_____CCCCCCUUUCAC___CUAU (((.....(((((((((.....((......))...)))))))))...)))..........((((.....................))))........................ (-15.12 = -15.00 + -0.12)


| Location | 13,928,973 – 13,929,073 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.25 |
| Mean single sequence MFE | -47.12 |
| Consensus MFE | -32.57 |
| Energy contribution | -36.20 |
| Covariance contribution | 3.63 |
| Combinations/Pair | 1.14 |
| Mean z-score | -4.10 |
| Structure conservation index | 0.69 |
| SVM decision value | 4.58 |
| SVM RNA-class probability | 0.999924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
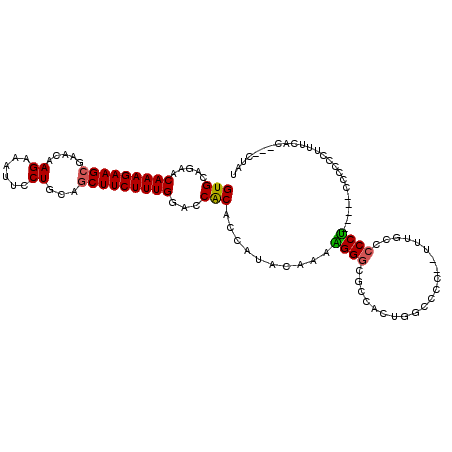
>X_DroMel_CAF1 13928973 100 + 22224390 ---GGG----GUUCCGUCCUUUUGUAUGGUGUGGUCCAAAGAAGCUGCAGGAGUUUCUUAUUUUCUUCUUUGUUCUGCACUCC-CAAAGG------------GAGGCCUUGGAUUUAUUA ---(((----(((...((((((((...((((..(..((((((((....(((.....))).....))))))))..)..))))..-))))))------------)))))))).......... ( -32.70) >DroSec_CAF1 9063 106 + 1 A--GGGGCCAGGGCCGCCCUUUUGUAUGGUGUGGUCCAAAGAAGCUGCAGGAAUUUCUUGUUCGCUUCUUUGUUCUGCACUCCCCAAAGG------------CAGGCCUUGGAUUUAUUA .--....(((((((((((..((((...((((..(..(((((((((.(((((.....)))))..)))))))))..)..))))...))))))------------).))))))))........ ( -50.20) >DroEre_CAF1 10816 119 + 1 GGGGGUGUCGGUGGCGCCCCUUUGUACGGUGCGGUCCAAAGAAGCUGCAGGAAUUUCUGGUUCGCUUCUUUGUUCCGCACUCC-CAAAGGGGACGACCCGGUGAGGCCUUGGAUAUAUUA .(((((.(((.(((((((((((((...(((((((..(((((((((.((.((.....)).))..)))))))))..)))))))..-)))))))).))..))).))).))))).......... ( -58.90) >DroYak_CAF1 11054 112 + 1 A--GGAGCAAGGGGCGCCCCUUUG-----UGUGGUCCAAAGAAGCUGCAGGAAUUUCUUGUUCGCUUCUUUGUUCUGCACUCC-CAAAGGGGGCGAAAUGGCGAGGCCUUGGAUUUAUUA (--((.((......((((((((((-----((..(..(((((((((.(((((.....)))))..)))))))))..)..)))...-..))))))))).....))....)))........... ( -46.70) >consensus A__GGGGCCAGGGCCGCCCCUUUGUAUGGUGUGGUCCAAAGAAGCUGCAGGAAUUUCUUGUUCGCUUCUUUGUUCUGCACUCC_CAAAGG____________GAGGCCUUGGAUUUAUUA .......((((((((.((((((((...(((((((..(((((((((.(((((.....)))))..)))))))))..)))))))...))))))))............))))))))........ (-32.57 = -36.20 + 3.63)


| Location | 13,928,973 – 13,929,073 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.25 |
| Mean single sequence MFE | -36.23 |
| Consensus MFE | -18.99 |
| Energy contribution | -21.05 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
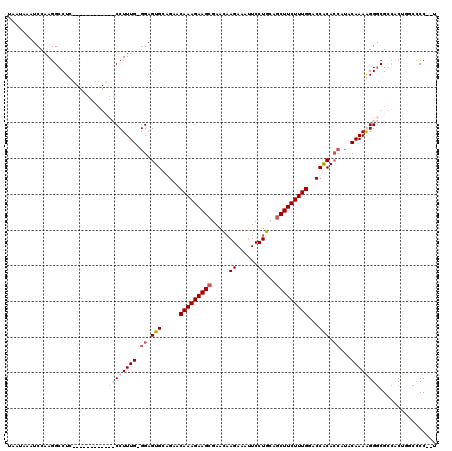
>X_DroMel_CAF1 13928973 100 - 22224390 UAAUAAAUCCAAGGCCUC------------CCUUUG-GGAGUGCAGAACAAAGAAGAAAAUAAGAAACUCCUGCAGCUUCUUUGGACCACACCAUACAAAAGGACGGAAC----CCC--- .......(((...(((((------------((...)-)))).))....((((((((......((......))....)))))))).......((........))..)))..----...--- ( -23.50) >DroSec_CAF1 9063 106 - 1 UAAUAAAUCCAAGGCCUG------------CCUUUGGGGAGUGCAGAACAAAGAAGCGAACAAGAAAUUCCUGCAGCUUCUUUGGACCACACCAUACAAAAGGGCGGCCCUGGCCCC--U ........(((.((((.(------------(((((..((.(((.....(((((((((.....((......))...)))))))))...))).))......)))))))))).)))....--. ( -37.10) >DroEre_CAF1 10816 119 - 1 UAAUAUAUCCAAGGCCUCACCGGGUCGUCCCCUUUG-GGAGUGCGGAACAAAGAAGCGAACCAGAAAUUCCUGCAGCUUCUUUGGACCGCACCGUACAAAGGGGCGCCACCGACACCCCC ............((..((....((.((.((((((((-...((((((..(((((((((....(((......)))..)))))))))..))))))....))))))))))))...))..))... ( -49.80) >DroYak_CAF1 11054 112 - 1 UAAUAAAUCCAAGGCCUCGCCAUUUCGCCCCCUUUG-GGAGUGCAGAACAAAGAAGCGAACAAGAAAUUCCUGCAGCUUCUUUGGACCACA-----CAAAGGGGCGCCCCUUGCUCC--U ............(((...))).....((((((((((-...(((.....(((((((((.....((......))...)))))))))...))).-----)))))))).))..........--. ( -34.50) >consensus UAAUAAAUCCAAGGCCUC____________CCUUUG_GGAGUGCAGAACAAAGAAGCGAACAAGAAAUUCCUGCAGCUUCUUUGGACCACACCAUACAAAAGGGCGCCACUGGCCCC__U ............................((((((((.((.(((.....(((((((((.....((......))...)))))))))...))).))...))))))))................ (-18.99 = -21.05 + 2.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:28 2006