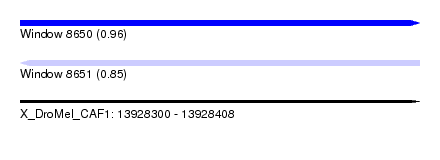
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,928,300 – 13,928,408 |
| Length | 108 |
| Max. P | 0.963116 |

| Location | 13,928,300 – 13,928,408 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.08 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -20.89 |
| Energy contribution | -20.73 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13928300 108 + 22224390 GCUGCAUUUGUGGGUCUUCCGCAUCCCUCUGAUUGGCCCCACAAGCUCCCCUAUA--UUC------UAUGUAUUCCUUUUUUUUUUUUUAAAGGAGUUUUGU-AGCUGC---AGCGGCAU ((((((((((((((....(((.(((.....)))))).))))))))..........--..(------((((.((((((((..........))))))))..)))-)).)))---)))..... ( -31.00) >DroSec_CAF1 8453 101 + 1 GCUGCAUUUGUGGGUCUUCCGCAUCCCUCGGAUUG-CCCCACAAACUCCCCUAUA--UUC------UAUAUAUUCCUCUUUUUU------AAGGAGUUUUGU-AGCUGC---AGCGGCAU ((((((((((((((.(.((((.......))))..)-.))))))))..........--..(------((((.((((((.......------.))))))..)))-)).)))---)))..... ( -33.80) >DroSim_CAF1 11335 101 + 1 GCUGCAUUUGUGGGUCUUCCGCAUCCCUCGGAUUG-CCCCACAAACUCCCCUAUA--UUC------UAUAUGCUCCUCUUUUUU------AAGGAGUUUUGU-AGCUGC---AGCGGCAU ((((((((((((((.(.((((.......))))..)-.))))))))..........--..(------((((.((((((.......------.))))))..)))-)).)))---)))..... ( -36.40) >DroEre_CAF1 10171 106 + 1 GCUGCAUUUGUGGGUCUUCCGCAGCCCGCUGAUUG-CCCCACAAACUCCCCCAUA--UUCCACAAGUACAUAUACA--UUUUUU------AAGGAGUUUUGUUAGCUGC---AGCGGCAU ((((((...(((((.(.....(((....)))...)-.)))))(((((((......--...................--......------..))))))).......)))---)))..... ( -27.01) >DroYak_CAF1 10051 104 + 1 GCUGCAUUUGUGGGUCUUCCGCAUCCCUCUGAUUG-CCCCACAAACUCCCCUAUAUAUUC------UAUAUAUUCC--AUUUUU------AAGGAGUUUUGU-AGCUGCAGCAGCGGCAU ((((((((((((((......(((((.....)).))-)))))))))..............(------((((.(((((--......------..)))))..)))-)).))))))........ ( -28.70) >consensus GCUGCAUUUGUGGGUCUUCCGCAUCCCUCUGAUUG_CCCCACAAACUCCCCUAUA__UUC______UAUAUAUUCCU_UUUUUU______AAGGAGUUUUGU_AGCUGC___AGCGGCAU (((((..(((((((...((...........)).....)))))))(((((...........................................)))))................))))).. (-20.89 = -20.73 + -0.16)

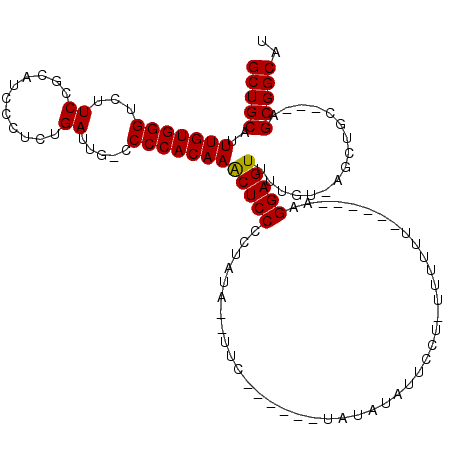
| Location | 13,928,300 – 13,928,408 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.08 |
| Mean single sequence MFE | -31.93 |
| Consensus MFE | -20.77 |
| Energy contribution | -21.01 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13928300 108 - 22224390 AUGCCGCU---GCAGCU-ACAAAACUCCUUUAAAAAAAAAAAAAGGAAUACAUA------GAA--UAUAGGGGAGCUUGUGGGGCCAAUCAGAGGGAUGCGGAAGACCCACAAAUGCAGC .....(((---(((...-......((((((((......................------...--..)))))))).(((((((.((.(((.....)))..))....))))))).)))))) ( -29.73) >DroSec_CAF1 8453 101 - 1 AUGCCGCU---GCAGCU-ACAAAACUCCUU------AAAAAAGAGGAAUAUAUA------GAA--UAUAGGGGAGUUUGUGGGG-CAAUCCGAGGGAUGCGGAAGACCCACAAAUGCAGC .....(((---(((...-...(((((((((------..................------...--....)))))))))(((((.-...((((.......))))...)))))...)))))) ( -31.11) >DroSim_CAF1 11335 101 - 1 AUGCCGCU---GCAGCU-ACAAAACUCCUU------AAAAAAGAGGAGCAUAUA------GAA--UAUAGGGGAGUUUGUGGGG-CAAUCCGAGGGAUGCGGAAGACCCACAAAUGCAGC .....(((---((..((-(.....((((((------......))))))....))------)..--.........(((((((((.-...((((.......))))...)))))))))))))) ( -33.70) >DroEre_CAF1 10171 106 - 1 AUGCCGCU---GCAGCUAACAAAACUCCUU------AAAAAA--UGUAUAUGUACUUGUGGAA--UAUGGGGGAGUUUGUGGGG-CAAUCAGCGGGCUGCGGAAGACCCACAAAUGCAGC ...(((((---(..(((.(((((.((((((------......--(.((((......)))).).--....)))))))))))..))-)...))))))((((((.............)))))) ( -33.62) >DroYak_CAF1 10051 104 - 1 AUGCCGCUGCUGCAGCU-ACAAAACUCCUU------AAAAAU--GGAAUAUAUA------GAAUAUAUAGGGGAGUUUGUGGGG-CAAUCAGAGGGAUGCGGAAGACCCACAAAUGCAGC .....((((((((..((-(((((.((((((------......--....((((((------...))))))))))))))))))).)-))......(((...(....).)))......))))) ( -31.50) >consensus AUGCCGCU___GCAGCU_ACAAAACUCCUU______AAAAAA_AGGAAUAUAUA______GAA__UAUAGGGGAGUUUGUGGGG_CAAUCAGAGGGAUGCGGAAGACCCACAAAUGCAGC .(((.(((.....)))........(((((.......................................))))).(((((((((....(((.....))).(....).)))))))))))).. (-20.77 = -21.01 + 0.24)

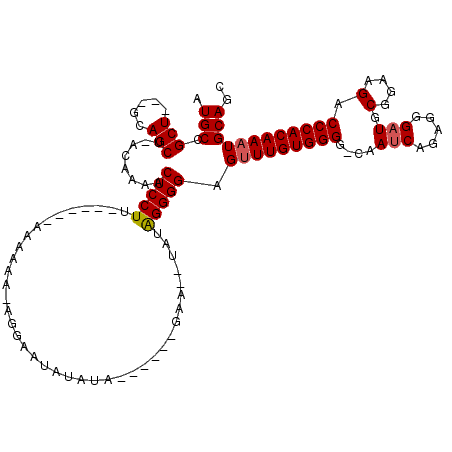

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:24 2006