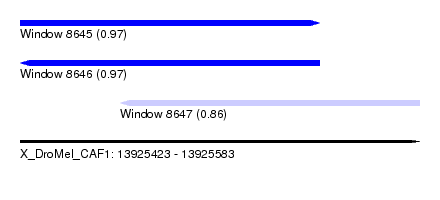
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,925,423 – 13,925,583 |
| Length | 160 |
| Max. P | 0.972360 |

| Location | 13,925,423 – 13,925,543 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -32.36 |
| Consensus MFE | -31.36 |
| Energy contribution | -31.96 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13925423 120 + 22224390 AAUUUCCGUGAAAUCUCCAAAUCAAACAAGACACAUGCGUUGCGCAUGCGCAUUGUCCACUGGCAUCUGCACCACGGCAACACUGCCACGCAAGUCCUGCCGCUACACUGAGCGAAAAGC ...(((.(((...................((((.((((((.......))))))))))....((((..........((((....))))((....))..))))....))).)))........ ( -28.80) >DroSec_CAF1 5594 120 + 1 AAUUUCCGUGAAAUCUCCAAAUCAAACAAGACACAUGCGUUGCGCAUGCGCAUUGUCCACUGGCAUCUGCACCACGGCAACACUGCCACGCAAGUCCUGCCGCUACACGGAGCGAAAAGC ...(((((((...................((((.((((((.......))))))))))....((((..........((((....))))((....))..))))....)))))))........ ( -34.90) >DroSim_CAF1 7538 120 + 1 AAUUUCCGUGAAAUCUCCAAAUCAAACAAGACACAUGCGUUGCGCAUGCGCAUUGUCCACUGGCAUCUGCACCACGGCAACACUGCCACGCAAGUCCUGCCGCUACACGGAGCGAAAAGC ...(((((((...................((((.((((((.......))))))))))....((((..........((((....))))((....))..))))....)))))))........ ( -34.90) >DroEre_CAF1 7474 120 + 1 AAUUUCCGUGAAAUCUUCAAAUCAAACAAGACACAUGCGUUGCGCAUGCGCAUUGUCCACUGGCAUCUGCAGCAUGGCAACACUGCCACGCAAGUCCUGCCGCUACACUGAGCGAAAAGC ...(((.(((...................((((.((((((.......))))))))))....((((.((...((.(((((....))))).)).))...))))....))).)))........ ( -31.00) >DroYak_CAF1 7066 120 + 1 AAUUUCCGUGAAAUCUCCAAAUCAAACAAGACACAUGCGUUGCGCAUGCGCAUUGUCCACUGGCAUCUGCACCACGGCAACACUGCCACGCAAGUCCUGCCGCUACACGGCGCGAAAAGU .....(((((...................((((.((((((.......))))))))))....((((..........((((....))))((....))..))))....))))).......... ( -32.20) >consensus AAUUUCCGUGAAAUCUCCAAAUCAAACAAGACACAUGCGUUGCGCAUGCGCAUUGUCCACUGGCAUCUGCACCACGGCAACACUGCCACGCAAGUCCUGCCGCUACACGGAGCGAAAAGC ...(((((((...................((((.((((((.......))))))))))....((((..........((((....))))((....))..))))....)))))))........ (-31.36 = -31.96 + 0.60)

| Location | 13,925,423 – 13,925,543 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -44.90 |
| Consensus MFE | -40.26 |
| Energy contribution | -40.66 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13925423 120 - 22224390 GCUUUUCGCUCAGUGUAGCGGCAGGACUUGCGUGGCAGUGUUGCCGUGGUGCAGAUGCCAGUGGACAAUGCGCAUGCGCAACGCAUGUGUCUUGUUUGAUUUGGAGAUUUCACGGAAAUU ..((((((....(..(.(((((((.(((.((...))))).))))))).)..).((..((((.((((((.(((((((((...))))))))).))))))...)))).....)).)))))).. ( -43.60) >DroSec_CAF1 5594 120 - 1 GCUUUUCGCUCCGUGUAGCGGCAGGACUUGCGUGGCAGUGUUGCCGUGGUGCAGAUGCCAGUGGACAAUGCGCAUGCGCAACGCAUGUGUCUUGUUUGAUUUGGAGAUUUCACGGAAAUU ((.....))((((((..(((((((.(((.((...))))).)))))))..........((((.((((((.(((((((((...))))))))).))))))...))))......)))))).... ( -45.80) >DroSim_CAF1 7538 120 - 1 GCUUUUCGCUCCGUGUAGCGGCAGGACUUGCGUGGCAGUGUUGCCGUGGUGCAGAUGCCAGUGGACAAUGCGCAUGCGCAACGCAUGUGUCUUGUUUGAUUUGGAGAUUUCACGGAAAUU ((.....))((((((..(((((((.(((.((...))))).)))))))..........((((.((((((.(((((((((...))))))))).))))))...))))......)))))).... ( -45.80) >DroEre_CAF1 7474 120 - 1 GCUUUUCGCUCAGUGUAGCGGCAGGACUUGCGUGGCAGUGUUGCCAUGCUGCAGAUGCCAGUGGACAAUGCGCAUGCGCAACGCAUGUGUCUUGUUUGAUUUGAAGAUUUCACGGAAAUU ..((((((((......)))((((......(((((((......)))))))......)))).((((((((.(((((((((...))))))))).))))).((((....)))).)))))))).. ( -42.60) >DroYak_CAF1 7066 120 - 1 ACUUUUCGCGCCGUGUAGCGGCAGGACUUGCGUGGCAGUGUUGCCGUGGUGCAGAUGCCAGUGGACAAUGCGCAUGCGCAACGCAUGUGUCUUGUUUGAUUUGGAGAUUUCACGGAAAUU ..((((((((((.....(((((((.(((.((...))))).)))))))))))).((..((((.((((((.(((((((((...))))))))).))))))...)))).....))..))))).. ( -46.70) >consensus GCUUUUCGCUCCGUGUAGCGGCAGGACUUGCGUGGCAGUGUUGCCGUGGUGCAGAUGCCAGUGGACAAUGCGCAUGCGCAACGCAUGUGUCUUGUUUGAUUUGGAGAUUUCACGGAAAUU ..((((((....(((..(((((((.(((.((...))))).)))))))..))).((..((((.((((((.(((((((((...))))))))).))))))...)))).....)).)))))).. (-40.26 = -40.66 + 0.40)

| Location | 13,925,463 – 13,925,583 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.98 |
| Mean single sequence MFE | -41.18 |
| Consensus MFE | -35.50 |
| Energy contribution | -36.30 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13925463 120 - 22224390 UAUACUUAUAAAUAUACCUAUCAAAGCGAGAAGAUGCCAAGCUUUUCGCUCAGUGUAGCGGCAGGACUUGCGUGGCAGUGUUGCCGUGGUGCAGAUGCCAGUGGACAAUGCGCAUGCGCA ........................(((((((((........)))))))))..((((((((((((.(((.((...))))).))))))).(((((..((((...)).)).))))).))))). ( -39.50) >DroSec_CAF1 5634 120 - 1 UAUACUCAUAAAUAUACCUGUCAAAGCGAGAAGAUGCCAAGCUUUUCGCUCCGUGUAGCGGCAGGACUUGCGUGGCAGUGUUGCCGUGGUGCAGAUGCCAGUGGACAAUGCGCAUGCGCA ................((((((..(((((((((........)))))))))..(.....)))))))...((((((((.(((((((((((((......)))).))).)))))))).)))))) ( -41.70) >DroSim_CAF1 7578 120 - 1 UAUACUCAUAAAUAUUCCUGUCAAAGCGAGAAGAUGCCAAGCUUUUCGCUCCGUGUAGCGGCAGGACUUGCGUGGCAGUGUUGCCGUGGUGCAGAUGCCAGUGGACAAUGCGCAUGCGCA ...............(((((((..(((((((((........)))))))))..(.....))))))))..((((((((.(((((((((((((......)))).))).)))))))).)))))) ( -42.70) >DroEre_CAF1 7514 108 - 1 UAUACC------------AUUCAUGGCGACAAGAUGCCAAGCUUUUCGCUCAGUGUAGCGGCAGGACUUGCGUGGCAGUGUUGCCAUGCUGCAGAUGCCAGUGGACAAUGCGCAUGCGCA ....((------------(((((((((((((...((((((((((.(((((......))))).))).))....))))).))))))))))..((....)).)))))....((((....)))) ( -39.80) >DroYak_CAF1 7106 118 - 1 UCUACUUAUAAAU--GCUUAUCAAAGCGACAAGAUGCCAAACUUUUCGCGCCGUGUAGCGGCAGGACUUGCGUGGCAGUGUUGCCGUGGUGCAGAUGCCAGUGGACAAUGCGCAUGCGCA ((((((..((..(--((..((((..((((((...(((((..(..(((..((((.....)))).)))...)..))))).))))))..)))))))..))..))))))...((((....)))) ( -42.20) >consensus UAUACUCAUAAAUAUACCUAUCAAAGCGAGAAGAUGCCAAGCUUUUCGCUCCGUGUAGCGGCAGGACUUGCGUGGCAGUGUUGCCGUGGUGCAGAUGCCAGUGGACAAUGCGCAUGCGCA .........................((((((((........))))))))...((((((((((((.(((.((...))))).))))))).(((((..((((...)).)).))))).))))). (-35.50 = -36.30 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:21 2006