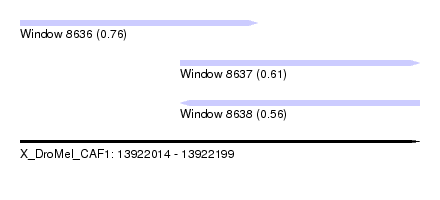
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,922,014 – 13,922,199 |
| Length | 185 |
| Max. P | 0.755743 |

| Location | 13,922,014 – 13,922,124 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.84 |
| Mean single sequence MFE | -36.64 |
| Consensus MFE | -12.16 |
| Energy contribution | -12.96 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
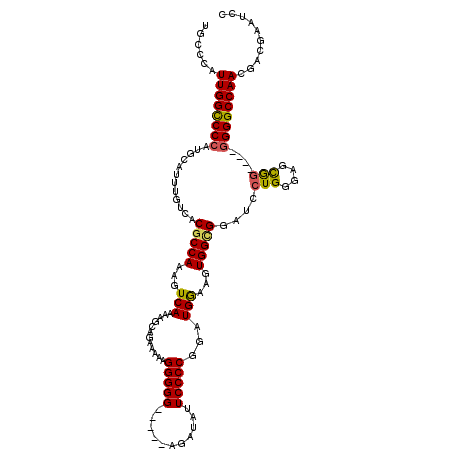
>X_DroMel_CAF1 13922014 110 + 22224390 AAA-AUGGUGGACUGGAAAGGAGGG-----GGUGGGGGUGGUGGGCGGGUUUUUGCGGAAAGAAUGUCAGUGUCUUUCAAGGAUUCGUCGUUGGCCCC----ACGCUCCCAGGAUCCGCC ...-..((((((((....))..(((-----(((((((((.(..(((((((((((...((((((.........)))))))))))))))))..).)))))----.)))))))....)))))) ( -48.40) >DroSec_CAF1 2230 89 + 1 -AA-AUGGUGGGUUGGGAACGUUGG-----GG--------------------CUGCGGAAAGAAUGUCAGUGUCUUUCAAGGAUUCGUCGUUGGCCCC----CUGCUCCCAGGAUCCGCC -..-..((((((((((((.((..((-----((--------------------((((((...((..(((..((.....))..))))).))).)))))))----.)).)))))..))))))) ( -35.10) >DroSim_CAF1 4227 89 + 1 -AA-AUGGUGGCUUGGAAACGUGGG-----GG--------------------CUGCGGAAAGAAUGUCAGUGUCUUUCAAGGAUUCGUCGUUGGCCCC----CUGCUCCCAGGAUCCGCC -..-..((((((((((...((.(((-----((--------------------((((((...((..(((..((.....))..))))).))).)))))))----)))...)))))..))))) ( -33.90) >DroEre_CAF1 3726 94 + 1 -AAAAGGGAGGGGUGAAAACAGGGGCAGGCGG--------------------UUGUGGAAAGAACGUCAGUGUCUUUCAAGGAUUCGUCGUUGGCCC-----CCACUCCCAGGCUCCACC -....(((((..((....)).(((((.(((((--------------------((.(.((((((((....)).)))))).).)).)))))....))))-----)..))))).......... ( -35.30) >DroYak_CAF1 3712 91 + 1 -AA-AUGGUGGGGUGCAAAGGGG-------GG--------------------UUGUGGAAAGAAUGUCAGUGUCUUUCAAGGAUUCGUCUUUGGCCCCAAACCCGCUGCCAGGAACCUCC -..-..((((((((.((((((.(-------((--------------------((.(.((((((.........)))))).).))))).)))))))))))....((.......)).)))... ( -30.50) >consensus _AA_AUGGUGGGGUGGAAACGGGGG_____GG____________________UUGCGGAAAGAAUGUCAGUGUCUUUCAAGGAUUCGUCGUUGGCCCC____CCGCUCCCAGGAUCCGCC ......((((((((...........................................((((((((....)).))))))............((((..............)))))))))))) (-12.16 = -12.96 + 0.80)

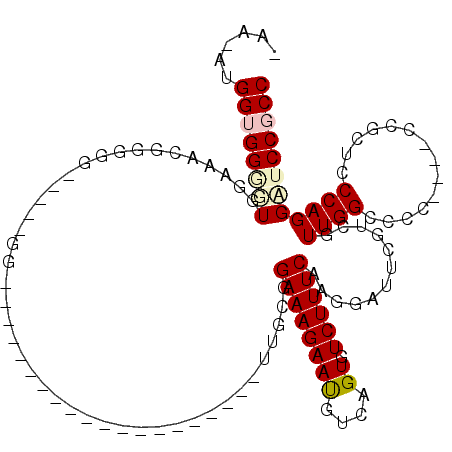
| Location | 13,922,088 – 13,922,199 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.23 |
| Mean single sequence MFE | -38.86 |
| Consensus MFE | -28.06 |
| Energy contribution | -28.86 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13922088 111 + 22224390 GGAUUCGUCGUUGGCCCC----ACGCUCCCAGGAUCCGCCACUUCCAUCCGGGGAAUAUCU-----CCCCCUUUUUCUGCUUUUGACUUUGGCGUGACAAAUGCAUGGGGCCAAUGGGCA ......((((((((((((----(.((((....))(((((((.........(((((.....)-----)))).....((.......))...))))).)).....)).)))))))))).))). ( -41.50) >DroSec_CAF1 2283 111 + 1 GGAUUCGUCGUUGGCCCC----CUGCUCCCAGGAUCCGCCACUUCCAUCCGGGGAAUAUCU-----CCCCCUUUUUCUGCUUUUGACUUUGGCGUGACAAAUGCAUGGGGCCAAUGGGCA ......((((((((((((----.(((((....))(((((((.........(((((.....)-----)))).....((.......))...))))).)).....))).))))))))).))). ( -41.50) >DroSim_CAF1 4280 111 + 1 GGAUUCGUCGUUGGCCCC----CUGCUCCCAGGAUCCGCCACUUCCAUCCGGGGAAUAUCU-----CCCCCUUUUUCUGCUUUUGACUUUGGCGUGACAAAUGCAUGGGGCCAAUGGGCA ......((((((((((((----.(((((....))(((((((.........(((((.....)-----)))).....((.......))...))))).)).....))).))))))))).))). ( -41.50) >DroEre_CAF1 3785 110 + 1 GGAUUCGUCGUUGGCCC-----CCACUCCCAGGCUCCACCACUUUCAUCCGGGGAAUAUGU-----CCCCCUUUUUCAGCUUCUGACUUUGGCGUGACAAAUGCAUGGGACCAAUGGGCA .............((((-----....(((((.((.....(((........(((((.....)-----))))........(((.........))))))......)).))))).....)))). ( -30.20) >DroYak_CAF1 3763 120 + 1 GGAUUCGUCUUUGGCCCCAAACCCGCUGCCAGGAACCUCCACUUUCAACCGGGGAAUAUAUAUCUCCCCCCUUUUUCUGCUGCUGACUUUGGCGUGACAAAUGCAUGGGGCCAAUGGGCA ......(((((((((((((.....((.((..((.....))..........(((((.........))))).........)).))........((((.....)))).))))))))).)))). ( -39.60) >consensus GGAUUCGUCGUUGGCCCC____CCGCUCCCAGGAUCCGCCACUUCCAUCCGGGGAAUAUCU_____CCCCCUUUUUCUGCUUUUGACUUUGGCGUGACAAAUGCAUGGGGCCAAUGGGCA ......((((((((((((......((.....((.....))..........((((.............)))).......))...........((((.....))))..))))))))).))). (-28.06 = -28.86 + 0.80)

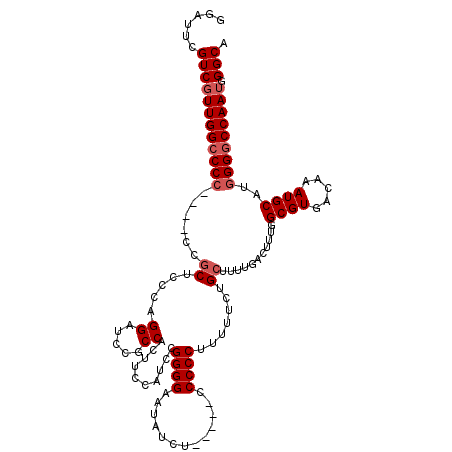

| Location | 13,922,088 – 13,922,199 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.23 |
| Mean single sequence MFE | -42.72 |
| Consensus MFE | -29.34 |
| Energy contribution | -29.02 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13922088 111 - 22224390 UGCCCAUUGGCCCCAUGCAUUUGUCACGCCAAAGUCAAAAGCAGAAAAAGGGGG-----AGAUAUUCCCCGGAUGGAAGUGGCGGAUCCUGGGAGCGU----GGGGCCAACGACGAAUCC ......((((((((((((.((((((((.(((..((.....)).......(((((-----(....))))))...)))..))))))))((....))))))----)))))))).......... ( -46.50) >DroSec_CAF1 2283 111 - 1 UGCCCAUUGGCCCCAUGCAUUUGUCACGCCAAAGUCAAAAGCAGAAAAAGGGGG-----AGAUAUUCCCCGGAUGGAAGUGGCGGAUCCUGGGAGCAG----GGGGCCAACGACGAAUCC ......((((((((.(((.((((((((.(((..((.....)).......(((((-----(....))))))...)))..))))))))((....))))).----)))))))).......... ( -42.60) >DroSim_CAF1 4280 111 - 1 UGCCCAUUGGCCCCAUGCAUUUGUCACGCCAAAGUCAAAAGCAGAAAAAGGGGG-----AGAUAUUCCCCGGAUGGAAGUGGCGGAUCCUGGGAGCAG----GGGGCCAACGACGAAUCC ......((((((((.(((.((((((((.(((..((.....)).......(((((-----(....))))))...)))..))))))))((....))))).----)))))))).......... ( -42.60) >DroEre_CAF1 3785 110 - 1 UGCCCAUUGGUCCCAUGCAUUUGUCACGCCAAAGUCAGAAGCUGAAAAAGGGGG-----ACAUAUUCCCCGGAUGAAAGUGGUGGAGCCUGGGAGUGG-----GGGCCAACGACGAAUCC .((((.(((.(((((.((.((..((((.((....((((...))))....(((((-----(....))))))))......))))..)))).))))).)))-----))))............. ( -39.70) >DroYak_CAF1 3763 120 - 1 UGCCCAUUGGCCCCAUGCAUUUGUCACGCCAAAGUCAGCAGCAGAAAAAGGGGGGAGAUAUAUAUUCCCCGGUUGAAAGUGGAGGUUCCUGGCAGCGGGUUUGGGGCCAAAGACGAAUCC ......(((((((((.((..(((((...(((...(((((..(.......)((((((........)))))).)))))...)))((....)))))))...)).))))))))).......... ( -42.20) >consensus UGCCCAUUGGCCCCAUGCAUUUGUCACGCCAAAGUCAAAAGCAGAAAAAGGGGG_____AGAUAUUCCCCGGAUGGAAGUGGCGGAUCCUGGGAGCGG____GGGGCCAACGACGAAUCC ......((((((((............(((((...(((............(((((...........)))))...)))...)))))....(((....)))....)))))))).......... (-29.34 = -29.02 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:13 2006